Figure 4.
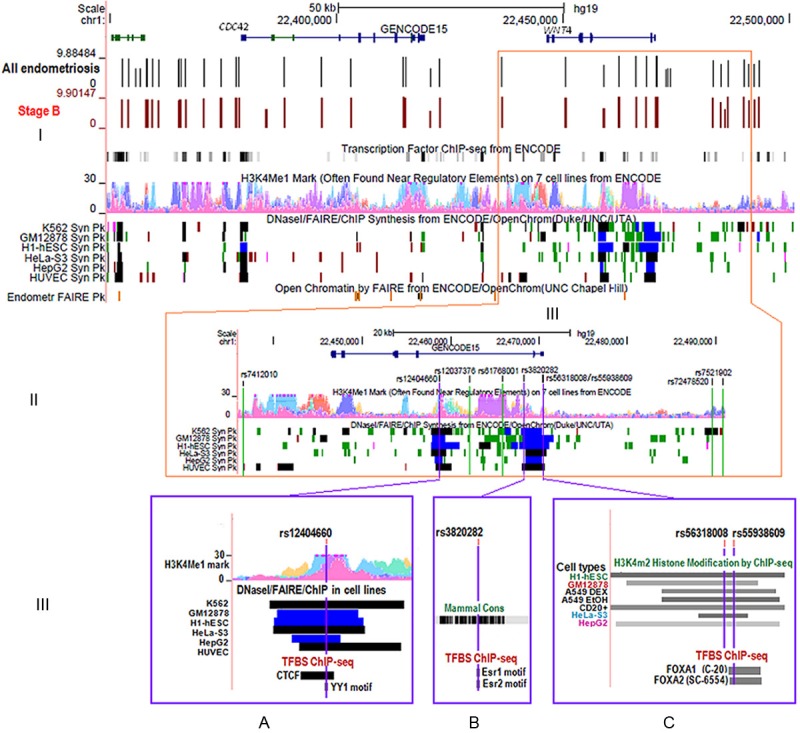
ENCODE annotation of WNT4 GWA and imputed variants in endometriosis. (I) Overview of the region of chromosome 1 including the CDC42 and WNT4 genes. Genotyped markers are shown by vertical lines with results (log-10 P values) for all endometriosis cases in black and stage B endometriosis cases in brown. Peaks for histone modification marks included in the ENCODE data [39] are shown for H3K4Me1 on 7 cell types (Tier 1 and Tier 2). The ENCODE data also shows areas of open chromatin as revealed by DNaseI, FAIRE and ChIP experiments in Tier 1 and Tier 2 cell types indicated by solid blocks (Regions showing peaks defined by both DNase I and FAIRE assays are in black, highly significant regions with combinations of peaks from these assays are in blue. Less significant regions identified by only DNase I HS as peaks are in green). The peak signals of open chromatin by FAIRE for normal endometrial tissue are shown in the Endometrium FAIRE Pk track. (II) Regions of interest around nine imputed SNPs. Three SNPs with the strongest association signals are located in regions of open chromatin as shown by DNaseI signals in the ENCODE data. (III) A zoomed-in image for the three top SNPs; (A) rs12404660, (B) rs3820282, and (C) rs55938609 (purple vertical lines) and functional annotation by ENCODE. Regulatory protein bound regions and histone modifications indicated by ChIP-seq of H3K4m2 in a subset of Tier 1 and 2 cell types (in greyscale light (lowest) to dark (highest)). The level of conservation across the region is shown for mammals. (A) SNP rs12404660 lies in histone modification area for H3K4Me1, open chromatin location by DNaseI, FAIRE, and ChIP assays, transcription factors CTCF by ChIP and sequence motifs of transcription factor YY1 by footprinting assays (B) rs3820282 is located within sequence motifs for transcription factors ESR1 and ESR2 and (C) rs55938609 lies within sequence bound by the transcription factors FOXA1 and FOXA2.
