Figure 1.
Expression of Lerboh1 and Wfi1 Transcripts and Rboh Polypeptide Levels in Antisense Plants.
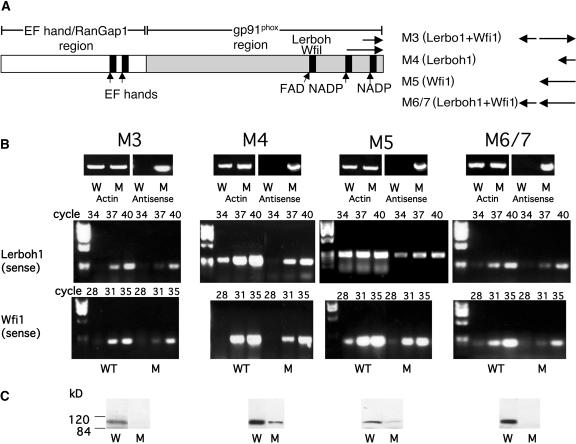
(A) Schematic presentation of L. esculentum Rboh showing gp91phox, EF hand, and RanGap homology regions. The location of the antisense fragments of Lerboh1 and Wfi1 are shown in the C-terminal region. The combinations of inserts used in constructs M3 to M6/7 are shown at the right. Fragment types are indicated in parenthesis, and the insert orientations are indicated at the right. Arrows pointing to the left or right represent antisense and sense orientations, respectively.
(B) RT-PCR expression analysis of antisense construct and Lerboh1 and Wfi1 sense expressions in wild-type (W) and antisense (M) plants. Top panels; RT-PCR product of antisense expression transcript showing fragments of 810, 293, 648, and 927 bp for M3, M4, M5, and M6/7, respectively; middle panels, RT-PCR of endogenous Lerboh1 sense transcript showing the expected PCR fragment of 270 bp; bottom panels, RT-PCR of endogenous Wfi1 sense transcript showing the expected PCR products of 213 bp. L. esculentum actin (Tom41 actin gene, U60480) was used as a standard, and the expected PCR product of a 325-bp fragment is shown. The data are representative of results obtained in at least four independent lines.
(C) Immunoblot analysis of L. esculentum Rboh levels in wild-type (W) and transgenic antisense (M) lines. Proteins were extracted from wild-type and antisense leaves and fractionated (100 μg per lane) by denaturating SDS-PAGE and immunoblotted with antisera against the C-terminal portion of the L. esculentum Rboh. The data are representative of results obtained in at least four independent lines.