Figure 1.
Isolation of T-DNA Insertions in the PP2AA2 and PP2AA3 Genes.
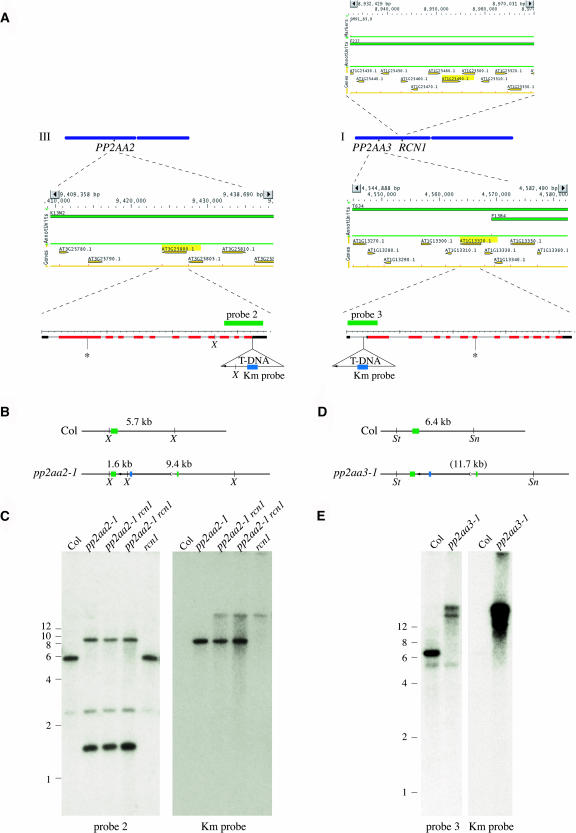
Chromosomes I and III are shown as blue bars (A), with the map positions of the RCN1 (At1g25490), PP2AA2 (At3g25800), and PP2AA3 (At1g13320) genes indicated. Map positions were determined using the Arabidopsis Information Resource chromosome map tool (http://www.arabidopsis.org/jsp/ChromosomeMap/tool.jsp). Above (RCN1) or below (PP2AA2 and PP2AA3) each chromosome is a MapViewer (http://www.arabidopsis.org/servlets/mapper) diagram of the chromosomal region surrounding the A subunit gene, which is highlighted in yellow. The structures of the PP2AA2 and PP2AA3 transcript splicing models, as shown in the Institute for Genomic Research database (http://www.tigr.org), are enlarged below, with exons (red bars), UTRs (black bars), and introns (lines) indicated for each. The gene structures are shown from 5′ (left) to 3′ (right), for example, in the opposite orientation from that shown in the MapViewer diagrams. The positions of the T-DNA insertions in pp2aa2-1 and pp2aa3-1 are shown, with the small arrowheads in each indicating the LB sequences. Asterisks indicate the positions of the pp2aa2-2 and pp2aa3-2 T-DNA insertions. Gene-specific probe fragments used in DNA gel blotting are shown as green bars, and the T-DNA–specific probe fragment (Km) is shown as a blue bar. The restriction map predicted for a single T-DNA insertion in the PP2AA2 locus (B) matches the DNA gel blotting data obtained with gene-specific (probe 2) and T-DNA–specific (Km) probes (C). The restriction map predicted for a single T-DNA insertion in the PP2AA3 locus (D) does not match the DNA gel blotting data (E). Genomic DNA was extracted from plants of the genotypes indicated and was digested with XbaI (C) or with SnaBI plus StuI (E). Weakly hybridizing bands were observed as a result of cross-hybridization of probe 2 with the PP2AA3 gene (C) and cross-hybridization of probe 3 with the PP2AA2 gene (E). The Km probe hybridizes with the rcn1 T-DNA (C). Sn, SnaBI; St, StuI; X, XbaI.