Figure 1.
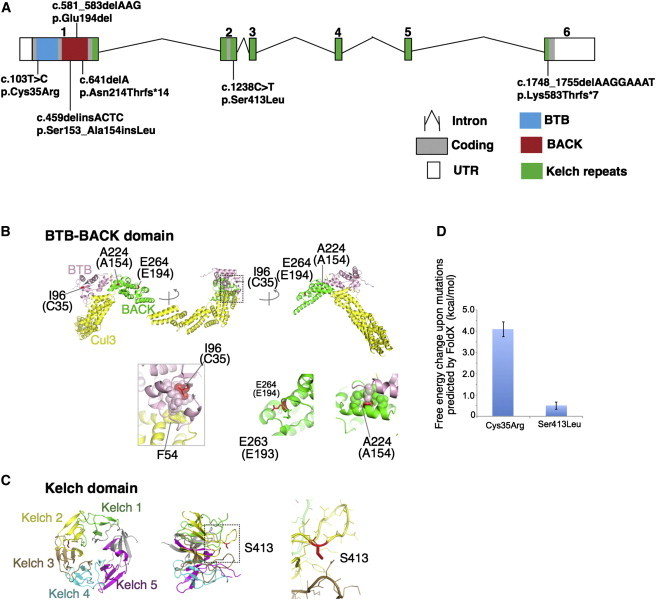
Overview of Mutations in KLHL41 and Their Effect on Protein Structure
(A) Schematic representation of mutations in KLHL41. Boxes represent exons 1–6. Conserved domains of KLHL41 are indicated as follows: BTB (blue), BACK (red), and Kelch repeats (green). The BTB and BACK domains are encoded by exon 1 and the five Kelch repeats are encoded by exons 1–6.
(B and C) Crystal structures of the BTB-BACK domain of human Kelch-like protein (KLHL11) in complex with CUL3 (Protein Data Bank code 4AP2) (B) and the Kelch domain of rat KLHL41 (PDB code 2WOZ) (C). α helices, β strands, and loops are drawn as ribbons, arrows, and threads, respectively. The squared areas correspond to the close-up views in the insets. In (B), the BTB and BACK domains are colored pink and green, respectively, whereas CUL3 is colored yellow, except that Ile96, Ala224, and Glu264 (Cys35, Ala154, and Glu194 in human KLHL41, respectively) are colored red. The side chains of these residues and Glu263 (Glu193 in human KLHL41) are shown as sticks with the indications of amino acid numbers for human KLHL11 and those for human KLHL41 in parentheses. Side chains involved in hydrophobic cores around Ile96 and Ala224 are drawn in van der Waal’s representation. In (C), the Kelch domain is color-coded to indicate each Kelch repeat, except that Ser413 is colored red. The side chain of Ser413 is shown as sticks. Molecular structures are drawn with PyMOL.
(D) Predicted free energy changes upon the substitutions of KLHL41 with FoldX software.
