Figure 1. Running time of different SW implementations for protein database search.
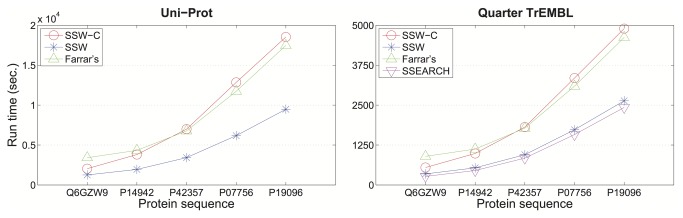
5 query proteins were searched against the whole Uni-Prot database (left) and one quarter of the TrEMBL database (right). Running time is shown on the y-axis for SSW without (blue) and with (red) detailed alignment, Farrar’s implementation (green) and SSEARCH (pink). All SW implementations used the BLOSUM50 scoring matrix with gap open penalty -12 and extension penalty -2.
