Figure 2. Running time of different SW implementations for simulated genomic read alignment.
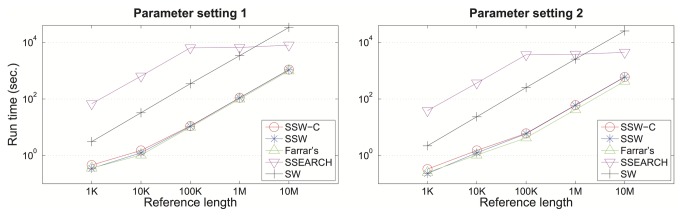
Running time of aligning 1,000 simulated Illumina reads to human reference sequences of various lengths. The log-scaled running time is shown on the y-axis for SSW without (blue) and with (red) detailed alignment, Farrar’s implementation (green), SSEARCH (pink) and an ordinary SW implementation (black). All SW implementations were tested under two sets of SW parameters: scores of match, mismatch, gap open and extension are 2, -1, -2, and -1 respectively (left), and scores of match, mismatch, gap open and extension are 1, -3, -5, and -2 respectively (right).
