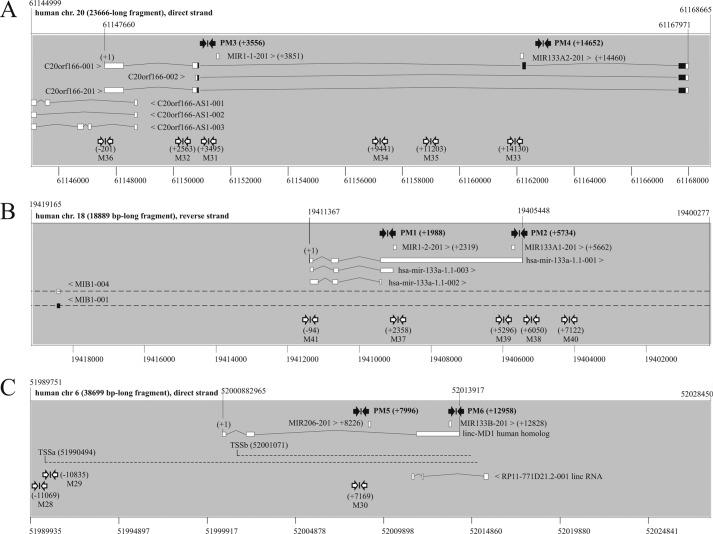
FIGURE 8.
Genes coding for human myogenic microRNAs miR-1, miR-133a, miR-133b, and miR-206. miRNA genes are transcribed into 5′-capped and 3′-polyadenylated primary microRNA precursors (pri-miRs) that are processed by Drosha/Pasha endonucleases into pre-miRs, which are exported to cytoplasm, where the final Dicer-dependent processing step and mature miRNA production take place (for a review, see Ref. 44). Positions of PCR primers used for pri-miR expression analysis via qRT-PCR and chromatin structure analysis via ChIP are indicated with black and white arrows, respectively. Human miR-1 and miR-133a are encoded within two common bicistronic genes on chromosomes 20 and 18. A, intron-exon structure and genome context (Ensembl) of the putative pri-miR precursor of miR-1-1 and miR-133a-2 on chromosome 20. The transcription start site (TSS) of this precursor is defined by the c20orf166 TSS according to CAGE tags, TSS seq, and H3K4me3 ChIP data (60). B, intron-exon structure and genome context (Ensembl) of the putative pri-microRNA precursor of miR-1-2 and miR-133a-1 on chromosome 18. The TSS of this precursor is given according to Ref. 60. The structure of the human miR-206 and -133b genes is currently unknown; however, it has been suggested that it might be similar to that of the corresponding mouse genes (61). In mice, miR-133b is expressed as a common precursor containing both miR-206 and miR-133b; miR-206, however, is expressed from its own internal promoter (61). C, putative intron-exon structure and genome context (Ensembl) of primary microRNA precursor of miR-206 and miR-133b on chromosome 6 deduced from its mouse homolog linc-MD1 (61); genomic coordinates are given according to genome assembly hg19; TSSa is according to Ref. 62; TSSb is according to Ref. 60. Genomic coordinates are given according to human genome assembly hg19. Pre-miRNA coordinates were extracted from Ensembl. Genomic coordinates of pri-miRNA precursors, including their putative TSSs, were extracted from the Refs. 60, 62, and 63. Conversion of genomic coordinates, if needed, was performed via the UCSC Genome Browser LiftOver utility.