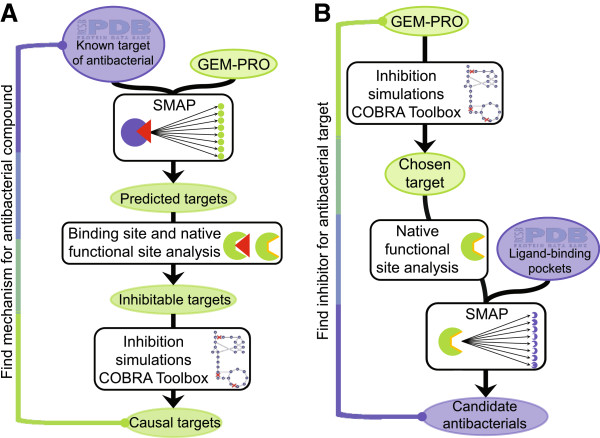
Figure 2.
Antibacterial prediction pipelines. (A) Screening causal targets for antibacterial activity of input compounds. Seeded with at least one structure of the compound of interest bound to a known target and the GEM-PRO to represent the functional proteome, SMAP is run to predict binding partners within the GEM-PRO. The potential for these predicted binding events to inhibit protein activity is then evaluated based on binding site overlap with native functional sites annotated in the GEM-PRO. Targets exhibiting overlap of antibacterial binding sites and functional sites are then evaluated for their inhibition growth phenotype in the GEM-PRO using the COBRA Toolbox. The inhibitable protein targets leading to deleterious growth phenotypes comprise predictions of causal targets for antibacterial activity. (B) Screening inhibitors of desired antibacterial target protein(s). Seeded with the GEM-PRO, metabolic simulations may be performed using the COBRA Toolbox to predict phenotypic impacts of protein inhibition to identify potential antibacterial target protein(s); alternatively, desirable targets may be chosen based on experimental results, such as gene-knockout phenotypes. To search for inhibitors of the chosen targets, the native functional sites of the proteins are identified, as in the GEM-PRO, and passed to SMAP to screen ligand-binding pockets of structures included in the PDB, searching for significant local structural matches. Significant matches comprise potential inhibitors of the chosen target proteins, expected to hold antibacterial properties.