Abstract
The relationship of body weight (BW) with white adipose tissue (WAT) mass and WAT gene expression pattern was investigated in mice submitted to physical training (PT). Adult male C57BL/6 mice were submitted to two 1.5-h daily swimming sessions (T, N = 18), 5 days/week for 4 weeks or maintained sedentary (S, N = 15). Citrate synthase activity increased significantly in the T group (P < 0.05). S mice had a substantial weight gain compared to T mice (4.06 ± 0.43 vs 0.38 ± 0.28 g, P < 0.01). WAT mass, adipocyte size, and the weights of gastrocnemius and soleus muscles, lung, kidney, and adrenal gland were not different. Liver and heart were larger and the spleen was smaller in T compared to S mice (P < 0.05). Food intake was higher in T than S mice (4.7 ± 0.2 vs 4.0 ± 0.3 g/animal, P < 0.05) but oxygen consumption at rest did not differ between groups. T animals showed higher serum leptin concentration compared to S animals (6.37 ± 0.5 vs 3.11 ± 0.12 ng/mL). WAT gene expression pattern obtained by transcription factor adipocyte determination and differentiation-dependent factor 1, fatty acid synthase, malic enzyme, hormone-sensitive lipase, adipocyte lipid binding protein, leptin, and adiponectin did not differ significantly between groups. Collectively, our results showed that PT prevents BW gain and maintains WAT mass due to an increase in food intake and unchanged resting metabolic rate. These responses are closely related to unchanged WAT gene expression patterns.
Keywords: Swimming training, Energy balance, White adipose tissue, WAT gene expression, C57BL/6 mice
Introduction
The metabolic properties of white adipose tissue (WAT) and its ability to produce hormones that are active in physiological processes and metabolic diseases have been investigated systematically (1). WAT-secreted adipokines such as resistin, tumor necrosis factor-alpha, interleukin 6, leptin, and adiponectin are associated with fat mass and may have effects on energy homeostasis, the immune system, reproductive function, and angiogenesis (2).
The plasticity of the adipose mass depends on the balance between the adipogenic, lipogenic and lipolytic activities of WAT. Adipocyte differentiation (adipogenesis) is mainly controlled by the transcription factor peroxisome proliferator-activated receptor-γ (PPAR-γ), which is activated by an agonist ligand in pre-adipocytes, promoting lipid accumulation and the expression of almost all genes characteristic of mature fat cells (3). Lipogenesis is the triacylglycerol biosynthesis and accumulation in the intracellular lipid droplet while lipolysis is the triacylglycerol hydrolysis (4). These metabolic processes respond to changes in energy balance and are associated with alterations in WAT mass and body weight (5).
The increase in WAT mass results from a positive balance between food intake and energy expenditure. Physical training has been known to improve energy expenditure and to induce a negative balance (6,7), to decrease WAT mass (8) and to ensure lower body weight (9). These responses can be associated with the increased lipolytic activity of WAT and the use of lipids as a source of energy (10). Moreover, the pre-adipocyte differentiation in mature adipocyte can be inhibited by physical training, resulting in decreased adipocyte number (11) and revealing that WAT mass modulation by physical training is associated with changes in size and number of adipocytes. However, the effect of physical training on adipokines is controversial. Some investigators have observed leptin reduction after physical training (12,13), while others have not (14). Adiponectin, an adipokine with action on insulin sensitivity and the inflammatory process (15), is increased after physical training associated with weight loss (13) or even with unaltered body weight, as shown by Hulver et al. (16).
Body weight, WAT mass and adipokine secretion associated with physical training have been studied. However, to our knowledge, the relationship between body weight, WAT mass and WAT gene expression pattern in mice submitted to physical training has not yet been investigated. Therefore, the purpose of the present study was to investigate the effects of physical training on body weight, WAT mass, WAT gene expression pattern obtained by the determination of the expression of transcription factor adipocyte and differentiation-dependent factor 1 (ADD1) involved in adipogenesis, fatty acid synthase (FAS) enzyme and malic enzyme expression as markers of lipogenesis, hormone-sensitive lipase (HSL) and adipocyte lipid binding protein (ALBP) expression as markers of lipolysis, and leptin and adiponectin gene expression.
Material and Methods
Mice
Ten-week-old male C57BL/6 mice were randomized into two groups, sedentary (S, N = 15) and exercise-trained (T, N = 18). Animals were maintained under the same housing conditions (12/12-h light/dark cycle and temperature 22°C), with free access to tap water and food. All experimental procedures were approved by the Research Ethics Committee of Escola de Educação Física e Esporte, Universidade de São Paulo (Protocol #25/2009).
Swimming training
The animals were submitted to physical training as described in detail by Evangelista et al. (17). Briefly, physical training consisted of two sessions of 1.5 h daily swimming, 5 days/week, for 4 weeks, without the addition of a workload. The swimming sessions initially lasted 20 min and were progressively increased by 10 min per day until reaching 1.5 h. This protocol was efficient in promoting aerobic adaptations such as resting bradycardia, cardiac hypertrophy and increased skeletal muscle oxidative capacity (17). In order to minimize the influence of water stress, sedentary mice were placed in the swimming apparatus for 5 min two times a week during the experimental protocol.
Body weight evolution
Body weight was measured weekly at the same time of day using a digital balance (Gehaka, Model BK4001, Brazil). Body weight gain was calculated as the difference between body weight measured at the beginning and at the end of the physical training protocol.
Metabolic measurements
Twenty-four-hour food intake was determined weekly throughout the study in the mice, which were kept 4 per cage. At the end of the physical training period, indirect calorimetry (Ametek, USA) after an overnight fast (8 h) was used to evaluate resting oxygen uptake per animal in a subgroup of mice randomly assigned to S (N = 6) and T (N = 6). The lowest amount of oxygen consumption observed for 30 min was considered to be the rest value. For the calculation of oxygen consumption, we used the following formula: VO2 = (FiO2 - FeO2) × 1165 / weight, where: FiO2 = oxygen inspired fraction, FeO2 = oxygen expired fraction; air pump flow = 1165 L.
Serum and tissue samples
Twenty-four hours after the end of the last training session, S and T mice were killed with an intravenous injection of pentobarbital sodium (4 mg/100 g body weight) and submitted to cardiac puncture. The tissues were harvested and the Lee index was determined by naso-anal length and body weight measurements (3√body weight/naso-anal length) (18). Blood was collected and centrifuged at 4°C (1000 g for 10 min) and serum was stored at -80°C in order to determine leptin concentration by radioimmunoassay using a species-specific antibody (kit from Linco® Research Inc., USA). The test sensitivity was 0.5 ng/mL leptin, and the intra- and interassay coefficients of variation were <5%.
Citrate synthase activity
Citrate synthase activity was determined as previously described by Alp et al. (19). Briefly, a sample of soleus muscle was homogenized for 30 s at maximum speed (Polytron PT 3100, Switzerland) in 300 µL extraction buffer (50 mM Tris-HCl and 1 mM EDTA, pH 7.4) and centrifuged (3800 g, 15 min, 4°C). Citrate synthase (EC 4.1.3.7) activity was measured by incubating 15 µL supernatant with assay buffer [100 mM Tris base, 0.2 mM 5,5′-dithiobis-(2-nitrobenzoic acid), 0.01% Triton X-100, 1 mg acetyl-CoA, pH 8.1]. The reaction was initiated with the addition of 10 µL 0.5 mM oxaloacetic acid to a final volume of 300 µL, and analyzed spectrophotometrically (412 nm) at 25°C. The protein content was quantified by the method of Bradford (20) using bovine serum albumin as a standard. Enzyme activity is reported as nmol·min−1·mg protein−1.
mRNA analysis by real-time PCR
Total RNA was extracted from periepididymal WAT using TRIzol®, according to manufacturer specifications (Invitrogen® Life Technologies, USA). In some cases, we pooled RNA from two mice because RNA obtained from the periepididymal WAT of one animal was insufficient. RNA samples (S, N = 10 and T, N = 10) were checked for integrity using agarose gel electrophoresis and were quantified spectrophotometrically at 260 and 280 nm in order to determine their concentration and extent of homogeneity. Only samples with a 260/280-nm ratio higher than 1.8 were utilized. SuperScript III reverse transcriptase (Invitrogen® Life Technologies) was used to reversely transcribe 2 µg total isolated RNA using an oligo(dT)10n primer (0.5 µg/µL, Invitrogen® Life Technologies) in a total reaction volume of 20 µL. The primers used for amplification of the studied genes are described in Table 1. The expression of cyclophilin was measured as an internal control for sample variation in the reverse transcriptase (RT) reaction.
Table 1. Primers used for gene amplification evaluated in white adipose tissue from sedentary and trained mice.
| Genes | Sequence |
|---|---|
| Leptin | F: 5′-AACCCTCATCAAGACCATTGTCA-3′ |
| R: 5′-CCTCTGCTTGGCGGATACC-3′ | |
| Adiponectin | F: 5′-CGGCAGCACTGGCAAGTT-3′ |
| R: 5′-CCGTGATGTGGTAAGAGAAGTAGTAGA-3′ | |
| ADD1 | F: 5′-GGAGCCATGGATTGCACATT-3′ |
| R: 5′-GGAAGTCACTGTCTTGGTTGTTGA-3′ | |
| FAS | F: 5′-TCCTGGAACGAGAACACGATCT-3′ |
| R: 5′-GAGACGTGTCACTCCTGGACTTG-3′ | |
| Malic enzyme | F: 5′-CTCCTTGTCCTTGGGCTCA-3′ |
| R: 5′-CACTACGCACAATCCGAGA-3′ | |
| HSL | F: 5′-TCTAGCATGGGGTCCAGAG-3′ |
| R: 5′-ACATGCCCCTCTACACGTC-3′ | |
| ALBP | F: 5′-GCGTGGAATTCGATGAAATCA-3′ |
| R: 5′-CCCGCCATCTAGGGTTATGA3′ | |
| Cyclophilin | F: 5′-AATGCGGACCAAACACAAA-3′ |
| R: 5′-CCTTCTTTCACCTTCCCAAA-3′ |
The primers were designed using the program Primer-Blast from NCBI-NIH (www.ncbi.nlm.nih.gov/tools/primer-blast/). F = forward; R = reverse; ADD1 = transcription factor adipocyte determination and differentiation-dependent factor 1; FAS = fatty acid synthase; HSL = hormone-sensitive lipase; ALBP = adipocyte lipid binding protein.
Gene expression was determined by real-time PCR and amplification was assessed with the ABI Prism 7700 Sequence Detection System (Applied Biosystems, USA) using the SYBR Green Master Mix reagent (Applied Biosystems). All samples were assayed in triplicate. The comparative threshold (CT) cycle method was used for data analysis. CT indicates the fractional cycle number at which the amount of amplified target reaches a fixed threshold, and ΔCT is the difference in threshold cycle for target and reference (cyclophilin). The levels of gene expression are given by 2−ΔΔCt, where ΔΔCT is the ΔCT value subtracted from the ΔCT of the control group (21).
Histological analysis
A retroperitoneal fat pad was used to determine adipocyte diameter. The tissue was embedded in Paraplast® (Structure Probe, Inc., USA), cut with a microtome and stained with hematoxylin-eosin. The diameter of 50 adipocytes per animal was measured using a computerized morphometric analysis system (Leica Quantimet 500, UK).
Statistical analysis
Data are reported as means ± SEM. The results were compared between the two groups using the Student t-test, except for body weight evolution, which was analyzed using one-way ANOVA for repeated measures. The Tukey post hoc test was used to determine differences between means when a significant change was observed with ANOVA. A P value of less than 0.05 was considered to be statistically significant.
Results
Body weight gain and adiposity in response to physical training
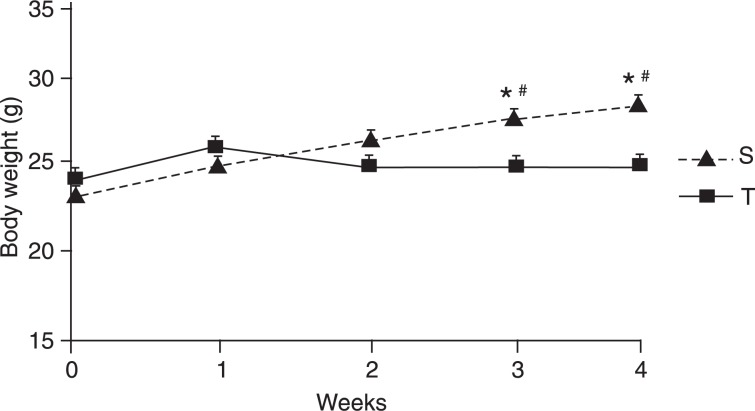
Changes in body weight during the 4 weeks of the experimental protocol are shown in Figure 1. No differences in body weight were observed between the S and T groups before physical training. However, S mice had a higher body weight than the T group during the 3rd and 4th weeks, also compared to their own body weight at the beginning of the experiment (P < 0.05). In fact, S animals had a substantial weight gain compared to T animals (4.06 ± 0.43 vs 0.38 ± 0.28 g, P < 0.01), confirming that physical training prevents body weight increases. In addition, the Lee index of S mice was significantly higher than that of T mice (30.4 ± 0.19 vs 29.3 ± 0.14 g/cm3, P < 0.05).
Figure 1. Body weight evolution during the experimental protocol in sedentary (S, N = 15) and trained (T, N = 18) mice. Data are reported as means ± SEM. Note that the S group had substantial weight gain compared to the T group. *P < 0.05 S vs T in the 3rd and 4th weeks. #P < 0.05 S in the 3rd and 4th weeks vs S in the 0 week (one-way ANOVA for repeated measures).
The weights of various internal organs and tissues were examined (Table 2). Even though the final body weight was lower in the T group, the weights of retroperitoneal and periepididymal fat pads, gastrocnemius and soleus muscle, lung, kidney, and adrenal glands did not differ significantly between groups. Liver and heart were larger in the T group while the spleen was smaller in the T group than in the S group (P < 0.05). Histological sections from the retroperitoneal fat pad of the S and T groups confirmed the absence of significant differences in adipocyte size (diameter) between groups (53.51 ± 2.4 vs 60.42 ± 2.7 µm, respectively).
Table 2. Weights of tissues and organs normalized for the total body weight of the animals.
| Tissues/organs | Sedentary (N = 15) | Trained (N = 18) |
|---|---|---|
| Retroperitoneal fat pad | 2.74 ± 1.48 | 2.76 ± 0.86 |
| Periepididymal fat pad | 11.75 ± 3.73 | 10.49 ± 2.77 |
| Gastrocnemius | 10.16 ± 0.83 | 9.82 ± 0.86 |
| Soleus | 0.56 ± 0.04 | 0.52 ± 0.05 |
| Lung | 7.04 ± 2.48 | 6.58 ± 0.62 |
| Kidney | 9.93 ± 0.65 | 10.39 ± 0.85 |
| Adrenal gland | 0.13 ± 0.02 | 0.15 ± 0.02 |
| Liver | 45.69 ± 4.62 | 49.57 ± 4.98* |
| Heart | 4.29 ± 0.22 | 4.99 ± 0.32* |
| Spleen | 4.03 ± 0.58 | 3.05 ± 0.45* |
Data (means ± SEM) are reported as mg/g body weight.
P < 0.05 compared to sedentary group (Student t-test).
Metabolic measurements
Food intake assessed over a period of 24 h was significantly higher in the T group during the physical training period when compared to the S group (4.7 ± 0.2 vs 4.0 ± 0.3 g·animal−1·24 h−1). There was no significant difference in oxygen consumption at rest between groups (S = 83.30 ± 2.9 and T = 79.66 ± 3.8 mL·kg−1·min−1). Serum leptin concentration was significantly higher in the T group compared to the S group (6.37 ± 0.5 vs 3.11 ± 0.12 ng/mL).
Gene expression in WAT
The levels of mRNA expression of the adipogenic transcription factor ADD1 and the enzymes for triacylglycerol synthesis, FAS and malic enzyme, were not influenced by physical training (Table 3). Similarly, the expression of the HSL and ALBP genes in the periepididymal fat pad, both markers of triglyceride hydrolysis, did not change after physical training. Despite the higher circulating concentrations of leptin in T mice, the expression level of leptin mRNA in periepididymal WAT assessed by real-time RT-PCR was similar for the S and T groups. Finally, no significant changes were observed in adiponectin gene expression (Table 3).
Table 3. mRNA levels of genes measured in the periepididymal white adipose tissue.
| Gene | Sedentary (N = 10) | Trained (N = 10) |
|---|---|---|
| ADD1 | 1.0 | 1.44 |
| FAS | 1.0 | 2.75 |
| Malic enzyme | 1.0 | 0.89 |
| ALBP | 1.0 | 1.18 |
| HSL | 1.0 | 1.75 |
| Leptin | 1.0 | 0.64 |
| Adiponectin | 1.0 | 2.53 |
Data are reported as arbitary units. ADD1 = transcription factor adipocyte determination and differentiation-dependent factor 1; FAS = fatty acid synthase; ALBP = adipocyte lipid binding protein; HSL = hormone-sensitive lipase. Data analyses were done by the comparative threshold (CT) cycle method and the levels of gene expression were given by 2−ΔΔCt, where ΔΔCT is the ΔCT (difference in threshold cycle for target and cyclophilin) value subtracted from ΔCT of the control group. The results were compared by the Student t-test (P > 0.05).
Citrate synthase activity
Physical training promoted a significant increase in the maximal activity of citrate synthase in the T group compared to the S group (148.6 ± 12.8 vs 208.1 ± 22.7 mmol·min−1·protein−1).
Discussion
The present study was conducted to determine the relationship between body weight, WAT mass and WAT gene expression pattern in mice submitted to physical training. Aerobic physical training has been used as an important tool to prevent overweight and obesity (8). The swimming protocol employed in the present study was found to be useful to promote an aerobic response on the basis of the increased activity of citrate synthase, an important hallmark of oxidative metabolism, observed in the T group (22).
The results of the present study showed body weight maintenance in the T group while the S group showed significant increases in body weight over the experimental period and an increased Lee index at the end of the protocol. Our body weight data corroborate previous studies that demonstrated body weight homeostasis in trained mice (22) and in girls subjected to physical training for 12 weeks (23).
However, the body weight response to physical training in animals is controversial in the literature. Benatti et al. (12) reported that healthy trained rats fed a chow diet showed a 10% reduction of body weight possibly due to the reduction of relative and absolute body fat, but this response is typically observed in trained animals previously submitted to a high-calorie diet (24,25) or with metabolic disorders (26) associated with impairment of the energy balance. In fact, a period of physical training has been shown to reverse the body weight gain induced by a cafeteria diet in rats and by a high-fat diet in mice, due to increased energy expenditure compared to sedentary animals (26,27).
The difference in body weight gain did not result in changes in WAT mass in S and T mice. In fact, in the present study, we found no effect of physical training on periepididymal and retroperitoneal mass or adipocyte size from the retroperitoneal fat pad. In view of the similar WAT mass in the presence of lower body weight, we measured most of the internal organs. Interestingly, the weight of muscle (gastrocnemius and soleus), lung, kidney, and adrenal glands did not differ between groups and only the liver and heart were larger and the spleen was smaller in the T group. It is important to note that WAT mass was determined by two main fat pads, but additional studies are necessary to explain the difference in body weight observed between S and T mice.
Considering that body weight is closely related to the energy balance (28), we studied some determinants of the latter. Food intake was significantly higher in the T group during the physical training period, indicating that an increased caloric intake is associated with physical training. In fact, Gollisch et al. (8) demonstrated that trained animals fed a normocaloric diet showed higher food intake (in grams) compared to the control group. Interestingly, Guerra et al. (29) observed that when food intake was similar in sedentary and trained rats, trained rats showed lower body weight and WAT mass than sedentary rats.
The resting metabolic rate assessed by indirect calorimetry was not altered by physical training in the present study. Although some determinants of daily energy expenditure were not evaluated in this study, such as energy expenditure during physical training and the thermogenic effect of food, and that the assessment of resting energy expenditure refers only to a period of 24 h, our results indicate that body weight maintenance in trained mice is due to increased caloric intake and to an unchanged resting metabolic rate.
In this study, the T group showed a significant increase in serum leptin concentration compared to the S group. This finding seems to be an acute effect of the last training session more than a chronic response because leptin was not able to modulate resting energy metabolism and to induce appetite inhibition in trained animals. Moreover, leptin gene expression, which is proportional to total body fat stores in rodents (30), did not differ between the two groups.
The ADD1 transcription factor has been shown to promote adipocyte differentiation by activating adipocyte-specific gene expression (31). The expression of ADD1 alone is capable of inducing modest adipogenesis (32). In the present study, physical training did not change ADD1 expression, suggesting that the adipogenesis process was not suppressed in the trained mice. Sakurai et al. (11) showed that physical training induced adipogenesis inhibition due to decreased PPARγ mRNA level in trained rats, which was associated with reduced body weight and WAT mass. The disagreement over the effects of physical training on adipogenesis might be due to the difference in energy balance, because Sakurai et al. (11) showed that the food intake of trained rats was not significantly different from that of sedentary rats, while in the present study we observed increased food intake in trained mice.
Given that the expression patterns in WAT can be altered by the state of energy balance (4), our gene expression results for trained mice are consistent with the food intake increase associated with physical training. In fact, the state of energy balance can induce changes in WAT gene expression and consequently modulate the body weight response. Lipogenic gene expression, such as FAS, malic enzyme and glucose-6-phosphate dehydrogenase is reduced during fasting and restored or overexpressed during refeeding (33). Petridou et al. (34) showed that physical training reduced FAS gene expression in periepididymal adipose tissue but, according to our results, this response disappeared when the energy balance was changed by increased food intake.
HSL is the predominant lipase effector of catecholamine-stimulated lipolysis in adipocytes (35). ALBP is potentially important for fatty acid handling proteins in coordinating lipid storage and mobilization, since it interacts directly with HSL and increases its activity and substrate availability (36). Although physical training exerts a potent stimulus increasing lipolysis, we did not observe a change in HSL or ALBP gene expression in the trained mice and, as a consequence, adipocyte size did not decrease with physical training.
Considering that leptin and adiponectin gene expression are influenced by fat mass (4), our study provided evidence that adipokine gene expression did not change when fat mass was maintained. In fact, some investigators have shown that the reduction of leptin and adiponectin becomes important when fat mass is reduced (37,38).
In conclusion, our data provide evidence that physical training prevented body weight gain and maintained WAT mass due to an increase in food intake and unchanged resting metabolic rate. These responses are closely related to unchanged WAT gene expression patterns. These results indicate that physical training may be a useful tool in the management of body weight, but the association of diet control may be important to obtain significant benefits regarding energy metabolism and WAT gene expression.
Acknowledgments
We thank V.S. Acauã for assistance with the citrate synthase activity technique and R.V. Mariliza for assistance with the gene expression technique. Research supported by PIBIC/CNPq to T.S. Higa and FAPESP (#09/52904-5 to F.S. Evangelista).
References
- 1.Cusi K. The role of adipose tissue and lipotoxicity in the pathogenesis of type 2 diabetes. Curr Diab Rep. 2010;10:306–315. doi: 10.1007/s11892-010-0122-6. [DOI] [PubMed] [Google Scholar]
- 2.Conde J, Scotece M, Gomez R, Gomez-Reino JJ, Lago F, Gualillo O. At the crossroad between immunity and metabolism: focus on leptin. Expert Rev Clin Immunol. 2010;6:801–808. doi: 10.1586/eci.10.48. [DOI] [PubMed] [Google Scholar]
- 3.Kim SP, Ha JM, Yun SJ, Kim EK, Chung SW, Hong KW, et al. Transcriptional activation of peroxisome proliferator-activated receptor-gamma requires activation of both protein kinase A and Akt during adipocyte differentiation. Biochem Biophys Res Commun. 2010;399:55–59. doi: 10.1016/j.bbrc.2010.07.038. [DOI] [PubMed] [Google Scholar]
- 4.Fonseca-Alaniz MH, Takada J, Alonso-Vale MI, Lima FB. [The adipose tissue as a regulatory center of the metabolism] Arq Bras Endocrinol Metabol. 2006;50:216–229. doi: 10.1590/s0004-27302006000200008. [DOI] [PubMed] [Google Scholar]
- 5.Ogasawara J, Nomura S, Rahman N, Sakurai T, Kizaki T, Izawa T, et al. Hormone-sensitive lipase is critical mediators of acute exercise-induced regulation of lipolysis in rat adipocytes. Biochem Biophys Res Commun. 2010;400:134–139. doi: 10.1016/j.bbrc.2010.08.026. [DOI] [PubMed] [Google Scholar]
- 6.Hunter GR, Brock DW, Byrne NM, Chandler-Laney PC, Del CP, Gower BA. Exercise training prevents regain of visceral fat for 1 year following weight loss. Obesity. 2010;18:690–695. doi: 10.1038/oby.2009.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Artham SM, Lavie CJ, Milani RV, Ventura HO. Value of weight reduction in patients with cardiovascular disease. Curr Treat Options Cardiovasc Med. 2010;12:21–35. doi: 10.1007/s11936-009-0056-y. [DOI] [PubMed] [Google Scholar]
- 8.Gollisch KS, Brandauer J, Jessen N, Toyoda T, Nayer A, Hirshman MF, et al. Effects of exercise training on subcutaneous and visceral adipose tissue in normal- and high-fat diet-fed rats. Am J Physiol Endocrinol Metab. 2009;297:E495–E504. doi: 10.1152/ajpendo.90424.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keith SW, Redden DT, Katzmarzyk PT, Boggiano MM, Hanlon EC, Benca RM, et al. Putative contributors to the secular increase in obesity: exploring the roads less traveled. Int J Obes. 2006;30:1585–1594. doi: 10.1038/sj.ijo.0803326. [DOI] [PubMed] [Google Scholar]
- 10.Lima-Silva AE, Adami F, Nakamura FY, Oliveira FR, Gevaerd MS. Fat metabolism during exercise: regulatory mechanisms. Braz J Human Performance Kinanthropol. 2006;8:106–114. [Google Scholar]
- 11.Sakurai T, Endo S, Hatano D, Ogasawara J, Kizaki T, Oh-ishi S, et al. Effects of exercise training on adipogenesis of stromal-vascular fraction cells in rat epididymal white adipose tissue. Acta Physiol. 2010;200:325–338. doi: 10.1111/j.1748-1708.2010.02159.x. [DOI] [PubMed] [Google Scholar]
- 12.Benatti FB, Polacow VO, Ribeiro SM, Gualano B, Coelho DF, Rogeri PS, et al. Swimming training down-regulates plasma leptin levels, but not adipose tissue ob mRNA expression. Braz J Med Biol Res. 2008;41:866–871. doi: 10.1590/s0100-879x2008001000006. [DOI] [PubMed] [Google Scholar]
- 13.Balducci S, Zanuso S, Nicolucci A, Fernando F, Cavallo S, Cardelli P, et al. Anti-inflammatory effect of exercise training in subjects with type 2 diabetes and the metabolic syndrome is dependent on exercise modalities and independent of weight loss. Nutr Metab Cardiovasc Dis. 2010;20:608–617. doi: 10.1016/j.numecd.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 14.Bouassida A, Chamari K, Zaouali M, Feki Y, Zbidi A, Tabka Z. Review on leptin and adiponectin responses and adaptations to acute and chronic exercise. Br J Sports Med. 2010;44:620–630. doi: 10.1136/bjsm.2008.046151. [DOI] [PubMed] [Google Scholar]
- 15.Simpson KA, Singh MA. Effects of exercise on adiponectin: a systematic review. Obesity. 2008;16:241–256. doi: 10.1038/oby.2007.53. [DOI] [PubMed] [Google Scholar]
- 16.Hulver MW, Zheng D, Tanner CJ, Houmard JA, Kraus WE, Slentz CA, et al. Adiponectin is not altered with exercise training despite enhanced insulin action. Am J Physiol Endocrinol Metab. 2002;283:E861–E865. doi: 10.1152/ajpendo.00150.2002. [DOI] [PubMed] [Google Scholar]
- 17.Evangelista FS, Brum PC, Krieger JE. Duration-controlled swimming exercise training induces cardiac hypertrophy in mice. Braz J Med Biol Res. 2003;36:1751–1759. doi: 10.1590/s0100-879x2003001200018. [DOI] [PubMed] [Google Scholar]
- 18.Bernardis LL, Patterson BD. Correlation between ‘Lee index’ and carcass fat content in weanling and adult female rats with hypothalamic lesions. J Endocrinol. 1968;40:527–528. doi: 10.1677/joe.0.0400527. [DOI] [PubMed] [Google Scholar]
- 19.Alp PR, Newsholme EA, Zammit VA. Activities of citrate synthase and NAD+-linked and NADP+-linked isocitrate dehydrogenase in muscle from vertebrates and invertebrates. Biochem J. 1976;154:689–700. doi: 10.1042/bj1540689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Evangelista FS, Krieger JE. Small gene effect and exercise training-induced cardiac hypertrophy in mice: an Ace gene dosage study. Physiol Genomics. 2006;27:231–236. doi: 10.1152/physiolgenomics.00022.2006. [DOI] [PubMed] [Google Scholar]
- 23.Nassis GP, Papantakou K, Skenderi K, Triandafillopoulou M, Kavouras SA, Yannakoulia M, et al. Aerobic exercise training improves insulin sensitivity without changes in body weight, body fat, adiponectin, and inflammatory markers in overweight and obese girls. Metabolism. 2005;54:1472–1479. doi: 10.1016/j.metabol.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 24.Vieira VJ, Valentine RJ, Wilund KR, Antao N, Baynard T, Woods JA. Effects of exercise and low-fat diet on adipose tissue inflammation and metabolic complications in obese mice. Am J Physiol Endocrinol Metab. 2009;296:E1164–E1171. doi: 10.1152/ajpendo.00054.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamashita AS, Lira FS, Rosa JC, Paulino EC, Brum PC, Negrao CE, et al. Depot-specific modulation of adipokine levels in rat adipose tissue by diet-induced obesity: the effect of aerobic training and energy restriction. Cytokine. 2010;52:168–174. doi: 10.1016/j.cyto.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Brandt N, De Bock K, Richter EA, Hespel P. Cafeteria dietinduced insulin resistance is not associated with decreased insulin signaling or AMPK activity and is alleviated by physical training in rats. Am J Physiol Endocrinol Metab. 2010;299:E215–E224. doi: 10.1152/ajpendo.00098.2010. [DOI] [PubMed] [Google Scholar]
- 27.Marques CM, Motta VF, Torres TS, Aguila MB, Mandarim-de-Lacerda CA. Beneficial effects of exercise training (treadmill) on insulin resistance and nonalcoholic fatty liver disease in high-fat fed C57BL/6 mice. Braz J Med Biol Res. 2010;43:467–475. doi: 10.1590/s0100-879x2010007500030. [DOI] [PubMed] [Google Scholar]
- 28.Muller MJ, Bosy-Westphal A, Later W, Haas V, Heller M. Functional body composition: insights into the regulation of energy metabolism and some clinical applications. Eur J Clin Nutr. 2009;63:1045–1056. doi: 10.1038/ejcn.2009.55. [DOI] [PubMed] [Google Scholar]
- 29.Guerra RL, Prado WL, Cheik NC, Viana FP, Botero JP, Vendramini RC, et al. Effects of 2 or 5 consecutive exercise days on adipocyte area and lipid parameters in Wistar rats. Lipids Health Dis. 2007;6:16. doi: 10.1186/1476-511X-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiraoka J, Hosoda K, Ogawa Y, Ikeda K, Nara Y, Masuzaki H, et al. Augmentation of obese (ob) gene expression and leptin secretion in obese spontaneously hypertensive rats (obese SHR or Koletsky rats) Biochem Biophys Res Commun. 1997;231:582–585. doi: 10.1006/bbrc.1997.6145. [DOI] [PubMed] [Google Scholar]
- 31.Gregoire FM, Smas CM, Sul HS. Understanding adipocyte differentiation. Physiol Rev. 1998;78:783–809. doi: 10.1152/physrev.1998.78.3.783. [DOI] [PubMed] [Google Scholar]
- 32.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertile F, Raclot T. mRNA levels of SREBP-1c do not coincide with the changes in adipose lipogenic gene expression. Biochem Biophys Res Commun. 2004;325:827–834. doi: 10.1016/j.bbrc.2004.10.110. [DOI] [PubMed] [Google Scholar]
- 34.Petridou A, Tsalouhidou S, Tsalis G, Schulz T, Michna H, Mougios V. Long-term exercise increases the DNA binding activity of peroxisome proliferator-activated receptor gamma in rat adipose tissue. Metabolism. 2007;56:1029–1036. doi: 10.1016/j.metabol.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 35.Miyoshi H, Souza SC, Zhang HH, Strissel KJ, Christoffolete MA, Kovsan J, et al. Perilipin promotes hormone-sensitive lipase-mediated adipocyte lipolysis via phosphorylation-dependent and -independent mechanisms. J Biol Chem. 2006;281:15837–15844. doi: 10.1074/jbc.M601097200. [DOI] [PubMed] [Google Scholar]
- 36.Shen WJ, Liang Y, Hong R, Patel S, Natu V, Sridhar K, et al. Characterization of the functional interaction of adipocyte lipid-binding protein with hormone-sensitive lipase. J Biol Chem. 2001;276:49443–49448. doi: 10.1074/jbc.M104095200. [DOI] [PubMed] [Google Scholar]
- 37.Miyazaki S, Izawa T, Ogasawara JE, Sakurai T, Nomura S, Kizaki T, et al. Effect of exercise training on adipocyte-size-dependent expression of leptin and adiponectin. Life Sci. 2010;86:691–698. doi: 10.1016/j.lfs.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 38.Kondo T, Kobayashi I, Murakami M. Effect of exercise on circulating adipokine levels in obese young women. Endocr J. 2006;53:189–195. doi: 10.1507/endocrj.53.189. [DOI] [PubMed] [Google Scholar]