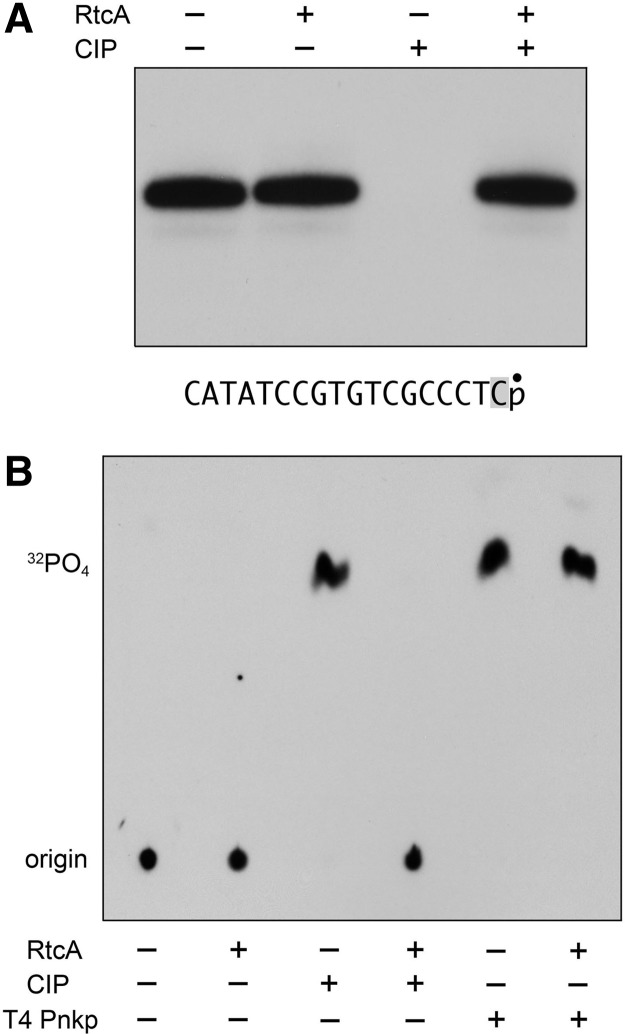
FIGURE 4.
Cyclization of DNA with a single 3′-terminal ribonucleotide. (A) Reaction mixtures (10 µL) containing 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 2 mM DTT, 100 µM ATP, 20 nM D17R13′p substrate (depicted at bottom with the 32P label denoted by • and the terminal ribonucleotide highlighted in gray), and either 400 nM RtcA (where indicated by +) or no added enzyme (−) were incubated for 30 min at 37°C. The mixtures were then treated with calf intestine phosphatase (CIP, where indicated by +) for 30 min at 37°C. The CIP-treated and mock-treated samples were mixed with an equal volume of formamide and EDTA, and then analyzed by electrophoresis through a 40-cm 20% polyacrylamide gel containing 7 M urea in 45 mM Tris borate, 1.2 mM EDTA. An autoradiogram of the gel is shown. (B) Reaction mixtures (10 µL) containing 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 2 mM DTT, 100 µM ATP, 0.2 µM D17R13′p substrate, and either 4 µM RtcA (where indicated by +) or no added enzyme (−) were incubated for 30 min at 37°C. The mixtures were then treated with either CIP or T4 polynucleotide kinase-phosphatase (Pnkp) for 30 min at 37°C. The treated and mock-treated samples were adjusted to 1 M formic acid and analyzed by PEI-cellulose TLC with 0.75 M LiCl as the mobile phase. An autoradiograph of the chromatogram is shown. The origin and the species corresponding to 32PO4 are indicated on the left.