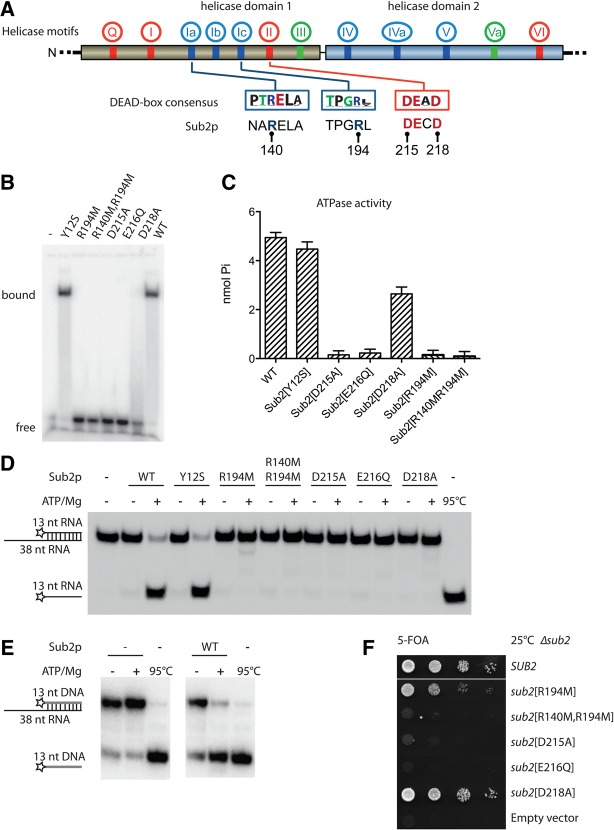
FIGURE 3.
Mutation of the NTM does not affect Sub2p helicase activity. (A) Schematic representation of mutations in the helicase core of Sub2p analyzed in this study. Location of the characteristic helicase motifs in DEAD-box proteins (red, motifs involved in ATP binding and hydrolysis; blue, motifs involved in RNA binding; green, motifs involved primarily in coupling between ATP and RNA binding). Boxes show the DEAD-box family consensus in motifs Ia, Ic, and II (Fairman-Williams et al. 2010); corresponding sequences of Sub2p are given underneath. The label for E216 is omitted for clarity. (B) In vitro RNA-binding gel-shift assay using a single-stranded 5′-radiolabeled 13-mer RNA. The migration of free and protein-bound RNA is shown. Sub2 was present at a concentration of 2 µM and the RNA at 0.6 nM. (C) ATPase activity measured by a modified Baginski-type assay (Cariani et al. 2004) using wt Sub2p or mutant variants as indicated and poly(U). The assay measures the relative production of Pi in nmol during a fixed time interval. (D) RNA-unwinding assay using a 13-bp duplex substrate with 12-bp overhang and wt or mutant Sub2p protein without (−) or with (+) ATP/Mg, as indicated. The positions of duplex substrate and unwound product are indicated at the left, where the stars show the position of the radiolabel. (E) RNA:DNA hybrid unwinding assay as in D but using a duplex of a 13-nt DNA annealed to a 38-nt RNA. (F) Tenfold serial dilution of yeast Δsub2 cells complemented by plasmids expressing wild type (SUB2), mutant Sub2p variants, or an empty vector control as indicated. Cells were plated on 5-FOA plates for 3 days at 25°C.