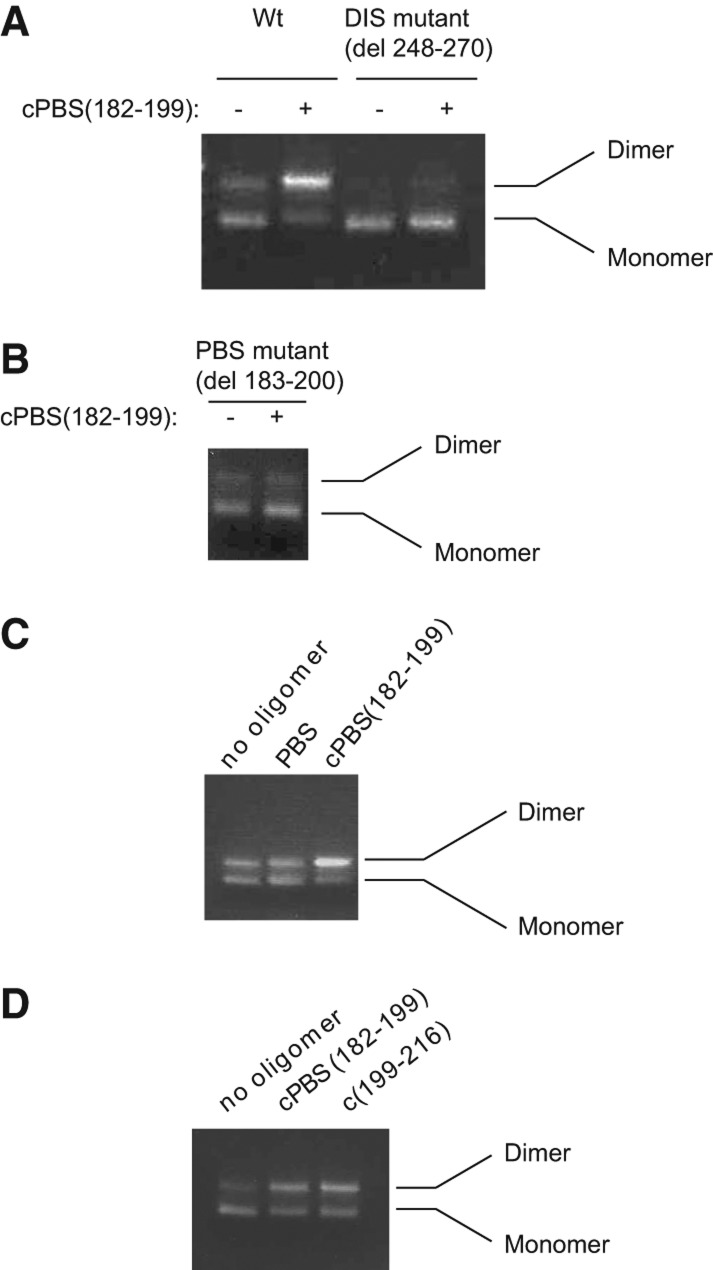
FIGURE 2.
Annealing to the PBS loop increases the dimerization of the 5′ UTR in a DIS dependent mechanism. (A) Resolution by agarose gel electrophoresis of the monomer and dimer of the 5′-UTR RNA, either wild-type or mutant RNA, ΔDIS, with nt 248–270 deleted (i.e., deletion of the DIS loop and part of the DIS stem). Gels are stained with ethidium bromide. The assays were performed as described in Materials and Methods in presence or absence of cPBS(182–199). (B) Dimerization of the 5′ UTR missing the PBS sequence, ΔPBS, in the presence or absence of cPBS(182–199). (C) Dimerization assay using wild-type 5′-UTR RNA that has been annealed with either a PBS oligomer, which has a sequence identical to the PBS sequence, or cPBS(182–199). This assay was performed in 200 nM 5′-UTR RNA. (D) Dimerization of the 5′ UTR annealed with either cPBS(182–199) or cPBS(199–216), oligomers complementary to two different regions within the PBS loop.