Abstract
Silver nanoparticles cause toxicity in exposed organisms and are an environmental health concern. The mechanisms of silver nanoparticle toxicity, however, remain unclear. We examined the effects of exposure to silver in nano-, bulk-, and ionic forms on zebrafish embryos (Danio rerio) using a Next Generation Sequencing approach in an Illumina platform (High-Throughput SuperSAGE). Significant alterations in gene expression were found for all treatments and many of the gene pathways affected, most notably those associated with oxidative phosphorylation and protein synthesis, overlapped strongly between the three treatments indicating similar mechanisms of toxicity for the three forms of silver studied. Changes in oxidative phosphorylation indicated a down-regulation of this pathway at 24 h of exposure, but with a recovery at 48 h. This finding was consistent with a dose-dependent decrease in oxygen consumption at 24 h, but not at 48 h, following exposure to silver ions. Overall, our data provide support for the hypothesis that the toxicity caused by silver nanoparticles is principally associated with bioavailable silver ions in exposed zebrafish embryos. These findings are important in the evaluation of the risk that silver particles may pose to exposed vertebrate organisms.
Introduction
Nanoparticles are being introduced, in some cases rapidly, into the consumer market but there is a fundamental lack of knowledge on their potential consequences for human and environmental health (reviewed in refs (1 and 2)). The use of silver nanoparticles (AgNP) has increased very significantly in recent years, with a current total global production estimated at 500 tonnes per year (reviewed in ref (1)). Very little empirical data are available on environmental levels of AgNP but given their very significant production, it is likely that AgNP are reaching the aquatic environment where they may impact on living organisms, as either particulates and/or as dissolved silver ions.
The susceptibility of freshwater fish to the toxic effects of exposure to silver ions (Ag+) is well documented and acute toxicity is associated predominantly with ionic Ag+ interactions at the gills, where they inhibit basolateral Na+/K+-ATPase activity. Inhibition of this enzyme compromises Na+ and Cl– uptake and therefore osmoregulation.3−6 The toxicity of this metal contaminant is affected by Ag speciation, which in turn is affected by a number of parameters including ionic composition and organic matter dissolved in the water. As a consequence of the decreased bioavailability of Ag with increased ionic strength of the water, fish inhabiting freshwater environments are more susceptible to Ag toxicity compared with marine fish species (reviewed in ref (6)).
A number of studies investigating the toxicity of AgNP, in a range of fish species, have provided evidence for concentration-dependent toxicity (reviewed in ref (1)). AgNP have been shown to enter zebrafish embryos via the chorion pore canals and were reported to be present in the brain, heart, yolk and blood of developing zebrafish.7,8 In the study by Asharani and colleagues,8 however, exposures to AgNP were conducted at very high concentrations and the particles had been stabilized with starch or BSA to prevent agglomeration. The conditions adopted were likely to enhance uptake in those laboratory exposures, but they may not reflect environmental reality. In juvenile or adult fish, one of the main routes of uptake and toxicity for AgNP appears to be via the gills, and adverse effects include on osmoregulation, similar to that reported for ionic silver.9,10 For AgNP, the effect on Na+/K+-ATPase activity in gills has been attributed to the dissolution of silver ions from the particles, and/or to the presence of silver ions at the surface of the AgNP.9 AgNP have also been reported to reduce the ability of fish to extract oxygen from the water during progressive oxygen depletion,11 without causing morphological alterations on the gill.9,11 The mechanisms by which this specific effect of exposure to AgNP occurs are unclear and may be due to damage of the gill surface caused by the presence of particulate matter and/or local dissociation of Ag+ from the AgNP trapped in the mucus surrounding the gill membrane. The biological processes affected by exposure to AgNP, reported across a variety of organs, include induction of oxidative damage, alterations to the regulation of enzymes responsible for free radical scavenging, altered regulation of gene expression pathways involved in apoptosis, and disrupted regulation of the cellular machinery involved in storing, detoxification and metabolism of metals (such as metallothionein 2).12−16
The objective of this study was to investigate the molecular mechanisms of toxicity of AgNP (10 nm) in comparison with a micrometer sized counterpart (Ag Bulk, 0.6-1.6 μm) and with Ag+, using a recently developed High-throughput (HT-)SuperSAGE approach on an Illumina GA II platform17 and zebrafish (embryos) as a model organism. Our data demonstrated that most transcriptomic alterations caused by exposure to AgNP and its ionic and bulk counterparts were common across all treatments, supporting the hypothesis that the toxicity of AgNP is principally associated with the toxicity of Ag+ at the surface of the particles or dissolved in the water. We also identified some gene changes unique to each treatment, suggesting that there may also be some particle-specific effects.
Materials and Methods
Characterization of the Silver Particles and Their Dissolution Rates
Silver particles (designated as AgNP [10 nm], and Ag Bulk [0.6–1.6 μm]) were purchased from Nanostructured and Amorphous Materials Inc., Houston, TX and silver with 2% HNO3 was obtained from PerkinElmer Life and Analytical Sciences, Shelton, CT. Further details on the characterization of these particles and their dissolution in exposure water can be found in ref (13) and in the Supporting Information (SI).
Fish Source, Culture, and Husbandry
Adult zebrafish (wild-type WIK strain, originally from the Max Planck Institute, Tubingen, Germany) for the provision of embryos were kept in aquaria at the University of Exeter in 140 L mixed-sex stock tanks, as described in ref (18). Fish were allowed to breed naturally and eggs were collected in glass egg chambers, approximately 1 h postfertilization (hpf). Eggs were then cleaned and unfertilized embryos were removed prior to the exposures.
Embryo Exposures to AgNP, Ag Bulk and Ag+ for Global Gene Expression Analysis
Stock solutions for AgNP, Ag Bulk and silver nitrate were made up in ultrapure water, and sonicated for 1 h to ensure dispersal of the particles. Exposures were conducted in glass chambers, at 28 ± 1 °C with a 12 h light:dark photoperiod. Immediately prior to the start of the exposures, glass chambers received 400 mL artificial water, prepared according to the ISO 7346–3:1996 guideline,19 containing 5 μg/L of 10 nm AgNP, 5 μg/L of Ag Bulk or 0.25 μg/L of silver nitrate. A control chamber was set up containing water alone. These exposure concentrations were well below those causing overt toxicity, based on the mortality curves for the various forms of silver obtained under the same experimental conditions in our laboratory (see SI Figure S1). The dosing of Ag Bulk was chosen to be the same as that for the AgNP (in terms of mass), to allow us to test whether the size of the particles was a determinant of the effects seen. An exposure to silver nitrate at 0.25 μg/L was included to provide a likely worst case scenario for the highest dissolution of Ag+ for the AgNP and Ag Bulk exposures in the adopted exposure conditions, based on data from the literature suggesting that dissolution of AgNP is variable but below 5% (Osborne et al 2012 reported this to vary between 0.1 and 2% in zebrafish embryo culture water (20)). Our own experiments (see SI) suggest that dissolution rates for AgNP and Ag Bulk under our experimental conditions were 0.27% ± 0.15% and 0.27% ± 0.07%, respectively. Exposures were initiated at 4 hpf and 585 ± 99 eggs were deployed into each treatment chamber. Unfertilized eggs were identified by visual inspection using light microscopy (Kyowa Optical SDZ PL, Kyowa Optical, Japan) and removed from the exposure chamber (22.8% of the total number of eggs were unfertilized). Solutions were replaced every 12 h during the exposure period, and dead embryos were removed at the times of replacement of the exposure water. At 24 h and 48 h, three pools of 50 embryos were removed from each of the exposure tanks, immediately frozen in liquid nitrogen and stored at −80 °C for analysis of gene expression. The experiment was terminated at 48 hpf.
Embryo Exposures for Investigations into the Effects of Ag+ on Oxygen Consumption in Zebrafish Embryos
In order to verify the biological significance of the effect of all treatments on the oxidative phosphorylation pathway, we designed a study to evaluate the effects of exposure to a series of doses of Ag+ on embryo oxygen consumption. Zebrafish embryos were exposed to a range of concentrations of Ag+ to include those expected to be present in the water following exposure to AgNP, Ag Bulk and Ag+ (0, 0.031, 0.062, 0.125, 0.250, 0.500, 1, and 5 μg Ag+/L). Exposures were initiated at 4 hpf and maintained for 48 h, and the average oxygen consumption per embryo was determined for two 24 h periods (0–24 h and 24–48 h from the onset of the exposure). The full description of the methodology adopted for these procedures is given in the SI.
RNA Isolation and HT-SuperSAGE Library Construction
RNA extraction was conducted on each pool of 50 embryos using the RNAeasy kit, according to the manufacturer’s instructions (Qiagen, UK). The quality and concentration of the resulting RNA were determined using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE), and an Agilent Bioanalyser 2100 (Agilent Technologies, Inc., Santa Clara, CA). HT-SuperSAGE was conducted according to previously described methods17 and libraries were sequenced using an Illumina Genome Analyzer (GA) II platform. Further details are given in the SI.
Sequence Tag Preprocessing
In total, over 16.9 million 36-bp sequences were obtained and processed, using the SuperSAGE data analysis procedure presented in Figure 1. A selection of tools from the FAST-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) and our own custom Perl scripts were used, running under the NEBC Bio-Linux environment.21 The FASTQ/A Barcode splitter (FASTX-Toolkit) was used to separate the samples from each lane based on the 4 base barcodes. The barcodes were removed from the sequences using the FASTQ/A trimmer (FASTX-Toolkit), before converting the sequences to FASTA format using FASTQ-to-FASTA (FASTX-Toolkit). A Perl script (SI, File S1) was used to remove all remaining adapter sequences after the last occurrence of CATG (NlaIII restriction site, used for sequence tag preparation) in each of the sequences. FASTX_collapser (FASTX-Toolkit) was used to collapse the sequences and calculate the frequency of unique sequence tags (unitags) in each library.
Figure 1.
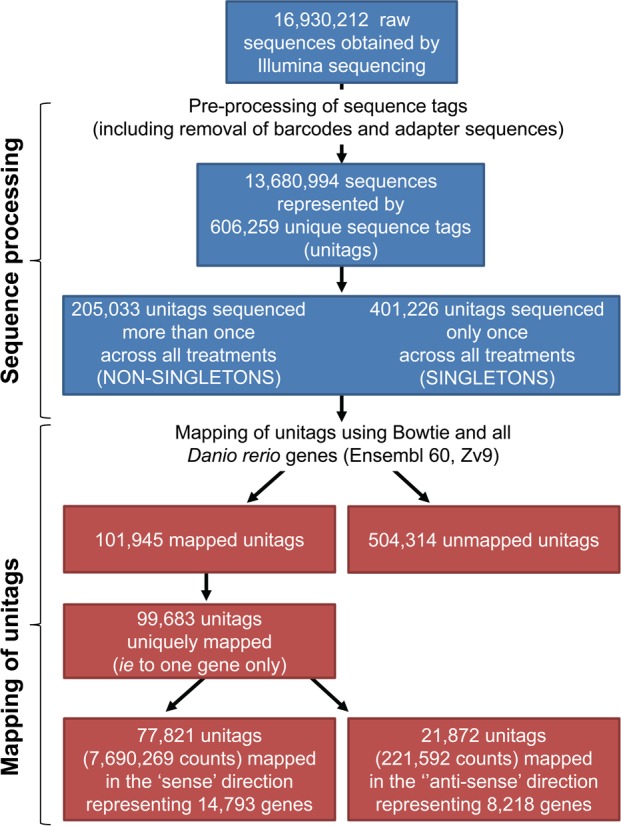
Flow diagram of the processing and annotation of the sequence tags.
Sequence Tag Mapping and Statistical Analyses
Unitags were mapped to all available zebrafish genes (28 491 genes) in Ensembl Zv9 (release 60; (22)) using Bowtie,23 with no mismatches allowed. Counts of unitags that mapped to the same gene were summed and unitags that aligned with more than one gene were removed from the data set using the Perl scripts provided in SI File S2. For each treatment, the counts of unitags representing each (unique) gene were normalized to 1 million tags. Fold changes (FC) were determined for each gene by dividing the number of tags in the normalized silver treatment libraries by the number of tags in the normalized control libraries. FC values less than 1 (down-regulated genes) were transformed using the following formula: −1/FC, in order to center the fold change values around 0. Differences in gene expression between the silver treatments and the controls were determined using DiscoverySpace 4.0,24 which implements the Audic-Claverie significance test to account for differences in sample size25 and P values were adjusted using a false discovery rate correction.26 Genes that were found to be significantly altered as a result of the silver treatments (adjusted P < 0.05) were further investigated. In order to visualize differences and similarities in gene expression between each of the silver treatments and the controls, scatter plots were produced using DiscoverySpace and correlation analysis (Pearson’s) was performed using Sigmaplot 11 (Systat Software UK Ltd.).
Functional Annotation of Differentially Expressed Genes
Functional annotation analysis was performed on the lists of differentially expressed genes (adjusted P < 0.05) for each treatment using the Database for Annotation, Visualization and Integrated Discovery (DAVID v6.7;27,28) and using all genes that were found to be expressed in zebrafish embryos at 24 hpf (12 939 genes) or 48 hpf (13 584 genes), respectively, as background. Gene Ontologies (GO) for Biological Processes, Cellular Components and Molecular Function were considered significantly enriched when P < 0.05.
Pathway Analysis
Pathway analysis was conducted through the use of Ingenuity Pathways Analysis (IPA; Ingenuity Systems, http://www.ingenuity.com), based on the lists of differentially expressed genes (adjusted P < 0.05) for each of the treatments. Enrichment of Canonical pathways was determined using the IPA library of canonical pathways, with a P value cutoff of 0.05 (adjusted for multiple testing using the Fisher’s Exact Test). Similarly, enriched KEGG pathways were determined using the lists of differentially expressed genes (adjusted P < 0.05) and all expressed genes at 24 hpf and 48 hpf, respectively, as background using DAVID.
Other Statistical Analysis
Statistical analysis of the data for embryo survival and oxygen consumption were conducted within SigmaPlot 11 (Systat Software UK Ltd.). Changes in embryo survival caused by exposure to AgNP, Ag Bulk or Ag+ during the exposure experiments for molecular analysis were analyzed using Chi-square tests. Relationships between the concentration of Ag+ and oxygen consumption were investigated using regression analysis. Differences between treatment groups were determined using one way analysis of variance (ANOVA), followed by an all pair-wise multiple comparison procedure (Student–Newman–Keuls Method). For these tests, data were considered to be significant when P < 0.05.
Data Deposition
The raw reads and processed unitag frequency tables for each of the HT-SuperSAGE libraries have been deposited in NCBI’s Gene Expression Omnibus29 and are accessible through GEO Series accession number GSE38125. (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE38125).
Results and Discussion
Embryo Mortality
Embryo mortality during the course of the exposures for generation of embryo samples for molecular analysis was 4.6 ± 2.5%, which falls within that expected for zebrafish and within the guidelines for OECD standardized testing guidelines.30 There were no differences in embryo mortality rates between treatment groups (P = 0.213), confirming that all exposures were conducted at concentrations below those causing any overt toxicity.
HT-SuperSAGE Data Analysis
The number of sequences obtained for each library ranged from 1.2 million (48 hpf AgNP) to 2.2 million (24 hpf AgNP) and the abundance distributions of these sequence tags were generally consistent between all libraries (SI Table S1). In total, 13 680 994 sequences were taken further for analysis and these were represented by 606 259 unique sequence tags (unitags). In total 99 683 unitags mapped to zebrafish genes, with 77 821 of these mapping to unique genes only (7 690 269 counts in total) and representing 14 793 genes (Figure 1). This is consistent with previous studies that reported a similar number of expressed genes in embryos at similar stages of development using zebrafish Affymetrix microarrays (ArrayExpress database [http://www.ebi.ac.uk/arrayexpress/]; Experiment ID: E-TABM-33) or RNA-Seq.31 Approximately 80% of the genes were represented by more than one unitag (see SI Figure S2), consistent with other studies that are based on SAGE methods, which is likely the result of incomplete NlaIII digestion during the library preparations, alternative polyadenylation and/or alternative splicing sites.32−34
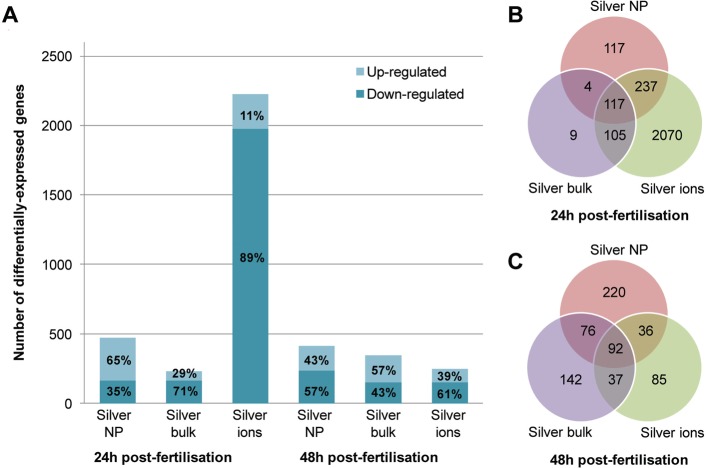
Statistical analysis of the gene expression data identified genes differentially expressed between the different treatments and the control, for both 24 hpf and 48 hpf (summarized in SI File S3). A visual representation of the expression profiles of each treatment group compared to the respective control is presented in SI Figure S3. There was a strong correlation between the levels of expression of genes in the controls compared with those in each treatment group (r > 0.937). The number of differentially expressed genes per treatment group, and the overlap between the differentially expressed genes for each treatment were consistent across all treatments, at both 24 h and 48 h of exposure (with the exception of the treatment 24 h Ag+; which showed a greater proportion of differentially expressed genes; Figures 2a, b, and c). A heatmap of the differentially expressed genes across the various silver treatment libraries is shown in SI Figure S4, and demonstrates the consistency in gene expression across all treatments. The condition tree highlights the differences in gene expression profiles for the differentially expressed genes between the 24 h Ag+ compared to all other treatments and between embryos collected at 24 hpf and 48 hpf.
Figure 2.
Differentially expressed genes in silver-treated zebrafish embryos. (a). The number of differentially expressed genes (adjusted P values <0.05) as a result of the various silver treatments (AgNP, Ag bulk and Ag+) compared to the controls. Percentages of up- and down-regulated genes for each of the silver treatments are indicated on the bars. (b, c). Venn-diagram showing the overlap between sets of differentially expressed genes obtained for each of the silver treatment libraries after 24 (b) and 48 (c) hours of exposure.
Some of the differentially expressed genes were found to be specific to each of the different treatments (AgNP – 13 genes; Ag bulk −1 gene; Ag+ - 32 genes), reflecting gene responses distinct to the different treatment (SI Table S2). Distinct genes were found to be differentially expressed at both time points for any one treatment and not present in the list of differentially expressed genes for the other treatments at any time point.
Commonalities in the Effects of Exposure to AgNP, Ag Bulk and Ionic Ag on Embryonic Gene Expression Profiles
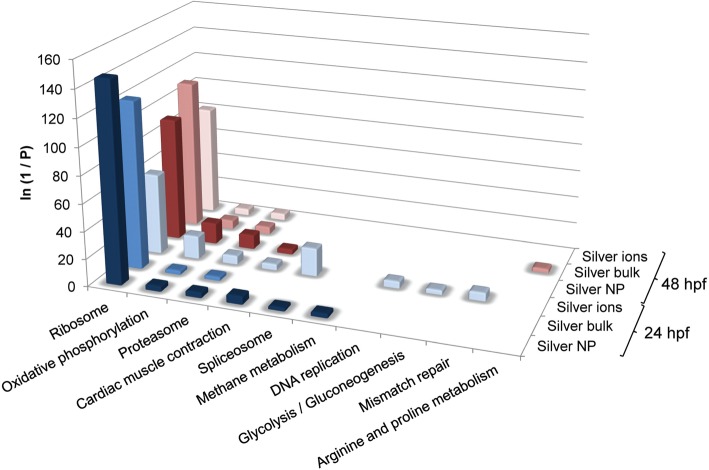
We explored the biological significance of the changes in gene expression caused by exposure to AgNP, Ag Bulk and Ag+ in 24 hpf and 48 hpf embryos by determining the enrichment in GO terms and canonical pathways using DAVID and IPA. Analyses were conducted based on differentially expressed genes, including all up- and down-regulated genes, to obtain a global overview of the biological effects of the three distinct treatments (Figure 3, SI Figure S5 and File S5). The most significantly over-represented GO terms and KEGG pathways (ribosome and oxidative phosphorylation) were common to all three treatments within each time of sampling, demonstrating a very significant overlap in the mechanisms of toxicity of the three forms or silver considered in this study. Based on this overlap, it seems likely that the toxicity of AgNP and Ag Bulk to zebrafish embryos is predominantly associated with the toxicity of free Ag+.
Figure 3.
KEGG pathways over-represented in differentially expressed gene lists following exposure of zebrafish embryos to silver. Over-representation analysis was conducted for gene lists differentially expressed between each treatment and its control (adjusted P values <0.05), after 24 and 48 h of exposure, using the list of all expressed genes as a background, within DAVID. The full data set for KEGG pathway over-representation is presented in SI, File S5.
The dissolution rates of AgNP and Ag Bulk in our experiments were 0.27% ± 0.15% and 0.27% ± 0.07%, respectively. Parallel studies in our laboratory, investigating the dissolution rates for AgNP and bulk silver in embryo culture water, have found them to vary between 0.1 and 2% over a 48 h period.20 Furthermore, very recently published data suggest that some silver particles (41.6 ± 9.1 nm AgNPs at a concentration of 0.2 nM) can penetrate the chorion through diffusion via the chorion canals35 and that once inside the chorion, the dissolution rate can be much higher compared to that observed in the exposure water.35 Consequently, the amount of bioavailable Ag+ in the AgNP and Ag Bulk treatments would likely be sufficient to cause most of the changes in gene expression observed. For many of the genes significantly affected in all the exposures, the changes were more pronounced (in terms of fold-change) for the exposure to Ag+ compared with those observed for AgNP and Ag Bulk, and this is likely to be associated, at least in part, with the bioavailability of silver ions in each treatment. However, the pathways affected (as assessed by the analysis of GO and KEGG pathway enrichment (Figure 3, SI Figure S5 and File S5)) remained common across all treatments. These changes are consistent with previous literature reporting effects of exposure to Ag (ions or nanoparticles) on several biological processes across a series of model organisms.9,36
The most significantly enriched GO terms for all treatments, both at 24 and 48 hpf were ribosome, translation and structural constituents of ribosome, for cellular components, molecular functions and biological processes, respectively. Concurrently, the most enriched KEGG pathway was ribosome (Figure 3 and SI File S5). These results align well with previous literature that report disruption to protein biosynthesis pathways in a range of organisms following exposure to AgNP and Ag+.37,38 Some of the most pronounced changes were observed for ribosomal proteins. For example, rpl10a was down-regulated by 5.6, 3.4 and 5.6-fold (24 h), and rpl18a down-regulated by 12.2, 1.2 and 9.7-fold (48 h), following exposure to AgNP, Ag Bulk, and Ag+, respectively. Expression of several genes encoding components of the ubiquitin proteasome system were also altered across all treatments, (at 24 and 48 h), indicating that protein degradation pathways were also altered by the treatments (SI File S3).
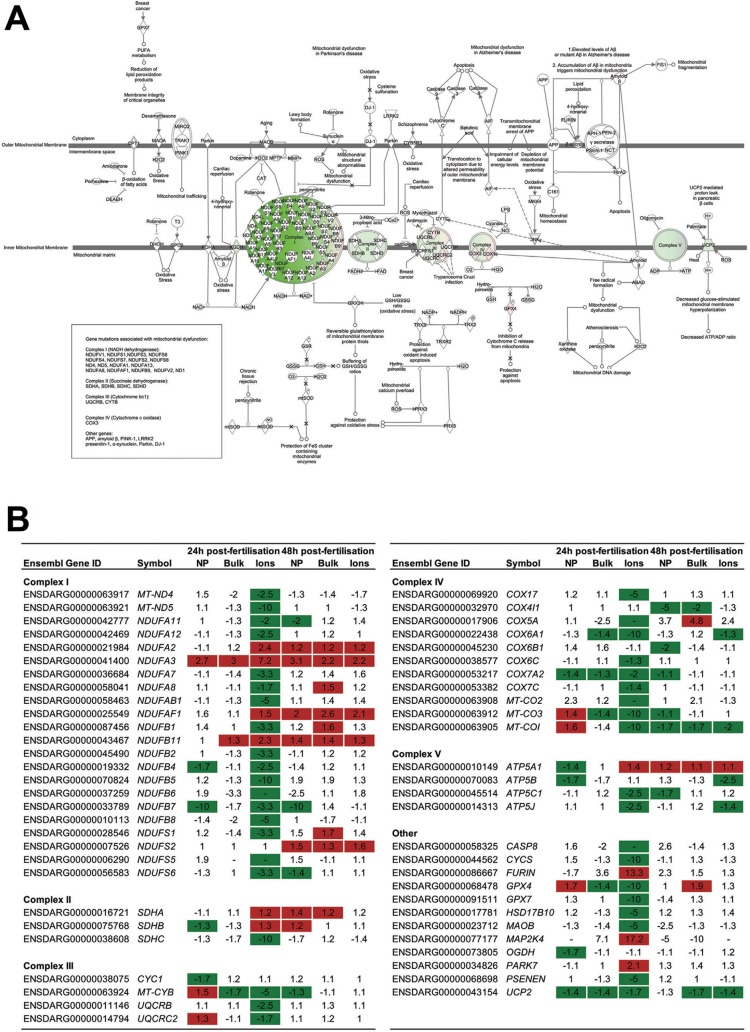
Oxidative phosphorylation was affected consistently by the exposure to AgNP, Ag Bulk, and Ag+ at both time points analyzed (Figure 3). This is a complex pathway that plays a key role in ATP generation in aerobic organisms, and is comprised of a series of five complexes and other associated molecules that act in concert within the mitochondrial inner membrane. The gene expression profiles following 24 h of exposure to all forms of Ag showed a down-regulation of many of the genes encoding for complex I, II, III and IV (Figure 4 and SI File S6). Within the complexes some genes were up-regulated whereas others were down-regulated, in particular for complex I (for Ag+ and Ag Bulk). This complex is comprised of a large number of genes that form 14 central and 32 accessory subunits, encoded by both the mitochondrial and the nuclear genome, and that associate into three functional modules, that oxidize NADH, reduce quinone and pump protons across the membrane (reviewed in ref (39)). Our data may reflect a differential regulation of the various subunits and/or a differential regulation of the various functions of complex I. Overall, following 24 h of exposure to all Ag treatments, the gene expression data indicated a down-regulation of all complexes involved in oxidative phosphorylation, possibly with the exception of complex I for Ag Bulk (Figure 4 and SI File S6). This effect is consistent with previously reported observations reporting a down-regulation of this pathway following Ag exposure in a human cell line, using biochemical methods.40 The degree to which these genes were regulated between the various treatments was consistent with predicted differences in the concentrations of bioavailable Ag+, which were expected to be highest for Ag+, followed by AgNP and lowest for Ag Bulk.
Figure 4.
Effects of the silver treatments on transcription of genes belonging to the mitochondrial dysfunction pathway. Shades of green and red represent down-regulation and up-regulation of target genes, respectively. (a) Effects of exposure to AgNP for 24 h on the mitochondrial dysfunction pathway. (b) Summary of the effects of exposure to all treatments on the mitochondrial dysfunction pathway for 24 h and 48 h. Numbers in the table represent fold-change compared to the time-matched control. Illustrations of the mitochondrial dysfunction and oxidative phosphorylation pathways for all six treatments are presented in SI, File S6. Pathways were generated through the use of Ingenuity Pathways Analysis (Ingenuity Systems, www.ingenuity.com).
Data for embryos analyzed following 48 h of exposure to all Ag treatments similarly showed significant alterations in the expression of genes encoding for proteins belonging to all complexes of the oxidative phosphorylation pathway. When taken together, the changes in gene expression suggest an up-regulation of these complexes, consistent with a recovery of the activity in this pathway, during the 24–48 h period, in exposed embryos (Figure 4 and SI File S5).
Quantifying Effects of Ag+ on Embryo Metabolism
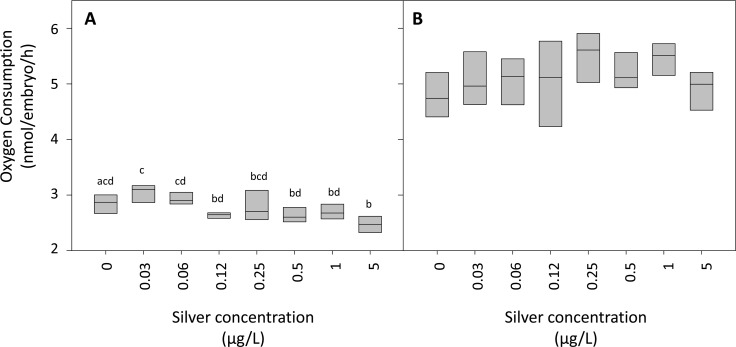
The changes in gene expression for constituents of the oxidative phosphorylation pathway suggested that Ag may cause a down-regulation in metabolic rates and ATP synthesis during the first 24 h of exposure, with a subsequent recovery from this effect at 48 h for all treatments. To further investigate the feasibility of this hypothesis, we conducted an experiment to investigate the effects of Ag+ on the metabolic activity, measuring oxygen consumption as a proxy for aerobic respiration of exposed embryos. Our data demonstrated that Ag+ caused a decrease in oxygen consumption during the first 24 h of exposure (r = −0.469; P < 0.001; Figure 5), with significant decreases in oxygen consumption in embryos exposed to 0.125, 0.5, and 5 μg Ag+/L compared with those exposed to 0.031 μg Ag+/L, and 5 μg Ag+/L compared to the controls during the first 24 h of exposure (One Way ANOVA followed by Student–Newman–Keuls Method; Figure 5a). This effect of Ag+ on embryo oxygen consumption was no longer apparent for the second 24 h period (between 24 and 48 h of exposure (r = −0.114; P = 0.467; Figure 5b), supporting the inhibitory effects of Ag+ on oxidative phosphorylation (and associated decrease in oxygen consumption) during the first 24 h of exposure and subsequent recovery by 48 h. Together, these data support previous studies that have reported inhibition of oxidative phosphorylation following exposure to silver nanoparticles and silver ions in a range of mammalian model systems.40−43 Importantly, our data suggest that zebrafish embryos have the capacity to deploy compensatory mechanisms at concentrations of Ag below those causing overt toxicity.
Figure 5.
Oxygen consumption rates of zebrafish embryos exposed to silver ions. Groups of 5 embryos were exposed to seven concentrations of silver ions (0–5 μg/L) in isolated glass vials, and the oxygen consumption was measured for 0–24h (A) and 24–48 h (B) exposure periods (n = six groups of five embryos per treatment). Average oxygen consumptions for individual embryos are presented. There was a significant effect of the exposure to silver ions for the 0–24 h, but not for the 24–48 h exposure periods (One Way ANOVA, P < 0.001, followed by an all pairwise comparison between the groups using the Student–Newman–Keuls Method, P < 0.05). Letters above the box plots represent statistical significance, with treatments with different letters being significantly different from each other.
The mechanisms by which exposure to Ag caused the observed changes in the oxidative phosphorylation pathway are difficult to ascertain. Recent evidence from mammalian studies has shown that pyruvate kinase, M2 (PKM2) that catalyzes the rate limiting ATP producing step of glycolysis, plays a key role in maintaining cellular redox homeostasis in cancer cells.44 This enzyme is specifically oxidized by H2O2, resulting in a decrease of its activity, decreased pyruvate formation and increased flux of glycolytic metabolites into the pentose phosphate pathway. This pathway produces reduced NADPH, a crucial source of reducing equivalents for fatty acid synthesis and for the glutathione, peroxiredoxin and thioredoxin systems, which play a key role in the detoxification of reactive oxygen species (ROS), providing cancer cells with a protective mechanism from intracellular ROS (reviewed in ref (45)). In contrast, normal mammalian cells express another pyruvate kinase isoform (PKM1), which is not susceptible to oxidation by hydrogen peroxide. Interestingly, we did not see expression of this later isoform in either 24 h or 48 h embryos. Instead, zebrafish embryos expressed two forms of the H2O2 sensitive isoform (pkm2a and pkmb2). The principal form of Pkm measured was pkm2a (the average counts per million in our data set were 226.0 ± 14.0 for embryos analyzed at 24 h and 565.5 ± 25.4 for embryos analyzed at 48 h). pkm2b was also expressed, but at much lower levels (0.9 ± 0.6 for 24 h embryos and 40.4 ± 8.4 for 48 h embryos). The fact that both the expressed forms of pkm in zebrafish are homologues to the human H2O2 sensitive form suggest that if the intracellular levels of H2O2 increase, zebrafish embryos may have the capacity to partially switch to the pentose phosphate pathway, as a mechanism of response to ROS. Increase in ROS is likely to occur during exposure to Ag, as demonstrated by the increase of gene markers for oxidative stress in our study, and in a number of other studies.46
One of the most significant changes in gene expression across the treatments was for alcohol dehydrogenase class-3 (adh5), which was 70.5, 7.1, and 125.0 fold up-regulated following exposure to AgNP, Ag Bulk, and Ag+, respectively, at 24 h. Similarly, after 48 h of exposure, adh5 was up-regulated by 151.0, 22.8, and 50.5, as a result of exposure to AgNP, Ag Bulk, and Ag+, respectively. Alcohol dehydrogenases (ADH) metabolize ethanol and other alcohols and aldehydes. In mammals, there are several classes of ADH, and class I ADH are responsible for the metabolism of ethanol and other small chain alcohols. Class II ADH enzymes preferentially metabolize larger alcohols and aldehydes (reviewed in ref (47)). In the zebrafish, enzymes structurally and functionally similar to the mammalian ADHs have been identified. However, the affinity of these enzymes to alcohols appears to be less specialized than that for mammalian enzymes48,49 but the affinity of each enzyme for the various alcohols has not been studied in detail. In addition to their role in alcohol metabolism, ADHs also convert NAD+ to NADH, and this may act as a compensatory mechanism for potential reduction in NADH resulting from the inhibition of the Pkm. The significance of the very pronounced increase in adh5 following exposure to all three forms of Ag is, therefore, difficult to ascertain. We hypothesize that this could be associated with the effects of all treatments on the oxidative phosphorylation pathway that may have resulted in a shift toward anaerobic metabolism, with increased accumulation of alcohol within the cells, and detoxification by Adh5.
Specific Effects of Exposure to AgNP, Ag Bulk and Ionic Ag on Embryonic Gene Expression Profiles
Analyses for differentially expressed genes across the different treatments and consistent over time (to investigate for possible particle related effects) identified 1 gene for Ag Bulk, 13 genes for the AgNP treatment and 32 genes for the Ag+ treatment, respectively (SI Table S2). These differences, however, would appear to be relative to the proportion of biologically available Ag+ in the different treatments and may not only indicate unique mechanisms associated with each treatment but also be a function of the level of bioavailable Ag+.
Considering the uniquely expressed genes in the different treatments, for Ag Bulk, ETS translocation variant 5 (etv5a) was 1.4 and 1.5-fold down-regulated at 24 h and 48 h, respectively. This gene is involved in the cellular response to oxidative stress (GO:0034599) and in the regulation of transcription, DNA-dependent (GO:00063550).
We identified 13 genes that were specific to AgNP. Among these genes, cryptochrome 1a (cry1a) was strongly down-regulated both at 24 and 48 h of exposure (6.3 and 4.3 fold down-regulated, respectively). This gene acts as a potent repressor of clock function and mimics the effect of constant light to “stop” the circadian oscillator, and is induced by light.50 This finding suggests that the light availability to embryos exposed to AgNP differed from that available to embryos in other treatments. Despite the fact that we tested relatively low concentrations of AgNP, it is possible that the suspended nanoparticles, and/or nanoparticles at the surface of the embryo, were acting as a barrier to light, and may have resulted in a difference in light availability to the embryos incubated in the AgNP treatment. Other genes responsive to AgNP alone included the proteasome subunit beta type-1 (psmb1), a gene involved in protein catabolism (1.2- fold down-regulated following 24h of exposure and 1.3-fold up-regulated at 48 h of exposure) and ribosomal protein S6 modification-like protein B (zgc:92164; 1.7-fold and 1.5- fold up-regulated at 24 h and 48 h, respectively). Together, these changes, although small in magnitude, indicate that the protein turnover in the AgNP treatment may be increased compared to the other treatments (in particular to Ag Bulk), suggesting that the potential damage caused by ROS on cellular proteins is compensated through degradation and synthesis of new proteins.
We identified 32 genes that were specific to the Ag+ treatment. The genes present in this list belong to gene ontologies that overlap with those over-represented among the genes affected by the other treatments (including oxidative phosphorylation and generation of precursor metabolites and energy). Genes regulated included Cyclic AMP-dependent transcription factor ATF-4 (atf4b1; 2.3 and 2-fold down-regulated at 24 h and 48 h, respectively); a gene involved in gluconeogenesis and regulation of transcription and probable ATP-dependent RNA helicase DDX56 (ddx56; 4.3 and 4.6-fold down-regulated at 24 h and 48 h, respectively), a gene involved in ribosome biogenesis. This adds to the evidence that the genes unique to this treatment reflect responses to higher levels of biologically available Ag+ associated with this treatment.
To summarize, we have applied sequencing-based transcription-profiling to investigate the mechanisms of toxicity of AgNP, compared to its ionic and bulk counterparts, in exposed zebrafish embryos, and provide mechanistic evidence for the hypothesis that the toxicity of AgNP is principally associated with the presence of bioavailable Ag+. All Ag treatments resulted in an inhibition of the oxidative phosphorylation pathway at 24 h of exposure, potentially due to the increase in ROS within mitochondria. Unique gene responses in the different treatments also supported that differences for responses in the transcriptome could be caused by the differences in bioavailability in Ag+ between the treatments. However, a gene response unique to AgNP was identified that may have resulted from alterations in light availability to the developing embryos, demonstrating that not all changes in gene expression observed can be explained by the effects of Ag+ alone, and that particle-associated effects may also contribute to the toxicity of AgNP.
Our work suggests that the predominant adverse effects of AgNP appear to be associated with the toxicity of Ag+. We would suggest that the risk assessments for AgNP should have a strong focus on understanding the bioavailability of Ag+ originating from AgNP, and the toxicology of Ag+, to establish what the levels of exposure and health consequences are likely to be in vulnerable environmental compartments.
Acknowledgments
We thank Dr. Matsumura for his support with the HT-SuperSAGE experiments, Dr. D. Studholme and Dr. D. Soanes for critical discussions on HT-SuperSAGE data analysis, Dr. C. Butler and Dr. M. van der Giezen for critical discussions on the biochemistry presented in this paper, Dr. R. Wilson for providing support for the oxygen consumption experiments and Dr Dogra for advice on the particle dissolution experiments. This work was funded by the Systems Biology Seed fund, University of Exeter to E.M.S., R.v.A. and C.R.T., and grants from the Natural Environment Research Council (NE/D004942/1 NE/C002369/1; NER/S/A/2005/13319 to C.R.T. and R.v.A.), the NERC Biomolecular Analysis Facility to T.B., and The UK Environment Agency to C.R.T.
Supporting Information Available
Further details on the methodology and results for the characterization of the silver particles used for the exposures, mortality curves, sequencing analysis, and a number of supporting figures and tables. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
⊥ B.D.J.: Comprehensive Pneumology Center, Institute of Lung Biology and Disease, Helmholtz Zentrum München, Ingolstädter Landstr. 1, 85764 Neuherberg, Germany
Author Contributions
∥ C.R.T. and E.M.S.: Joint Senior Authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Fabrega J.; Luoma S. N.; Tyler C. R.; Galloway T. S.; Lead J. R. Silver nanoparticles: Behaviour and effects in the aquatic environment. Environ. Int. 2011, 372517–31. [DOI] [PubMed] [Google Scholar]
- Scown T. M.; van Aerle R.; Tyler C. R. Review: Do engineered nanoparticles pose a significant threat to the aquatic environment?. Crit. Rev. Toxicol. 2010, 407653–70. [DOI] [PubMed] [Google Scholar]
- Hogstrand C.; Ferguson E. A.; Galvez F.; Shaw J. R.; Webb N. A.; Wood C. M. Physiology of acute silver toxicity in the starry flounder (Platichthys stellatus) in seawater. J. Comp. Physiol. B 1999, 1697461–73. [DOI] [PubMed] [Google Scholar]
- Bury N. R.; Wood C. M. Mechanism of branchial apical silver uptake by rainbow trout is via the proton-coupled Na(+) channel. Am. J. Physiol. 1999, 2775 Pt 2R1385–91. [DOI] [PubMed] [Google Scholar]
- Bury N. R.; Grosell M.; Grover A. K.; Wood C. M. ATP-dependent silver transport across the basolateral membrane of rainbow trout gills. Toxicol. Appl. Pharmacol. 1999, 15911–8. [DOI] [PubMed] [Google Scholar]
- Wood C. M.; Playle R. C.; Hogstrand C. Physiology and modeling of mechanisms of silver uptake and toxicity in fish. Environ. Toxicol. Chem. 1999, 18171–83. [Google Scholar]
- Lee K. J.; Nallathamby P. D.; Browning L. M.; Osgood C. J.; Xu X. H. In vivo imaging of transport and biocompatibility of single silver nanoparticles in early development of zebrafish embryos. ACS Nano 2007, 12133–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asharani P. V.; Lian Wu, Y.; Gong Z.; Valiyaveettil S. Toxicity of silver nanoparticles in zebrafish models. Nanotechnology 2008, 1925255102. [DOI] [PubMed] [Google Scholar]
- Griffitt R. J.; Hyndman K.; Denslow N. D.; Barber D. S. Comparison of molecular and histological changes in zebrafish gills exposed to metallic nanoparticles. Toxicol. Sci. 2009, 1072404–15. [DOI] [PubMed] [Google Scholar]
- Farmen E.; Mikkelsen H. N.; Evensen O.; Einset J.; Heier L. S.; Rosseland B. O.; Salbu B.; Tollefsen K. E.; Oughton D. H. Acute and sub-lethal effects in juvenile Atlantic salmon exposed to low mug/L concentrations of Ag nanoparticles. Aquat. Toxicol. 2012, 108, 78–84. [DOI] [PubMed] [Google Scholar]
- Bilberg K.; Malte H.; Wang T.; Baatrup E. Silver nanoparticles and silver nitrate cause respiratory stress in Eurasian perch (Perca fluviatilis). Aquat. Toxicol. 2011, 962159–65. [DOI] [PubMed] [Google Scholar]
- Roh J. Y.; Sim S. J.; Yi J.; Park K.; Chung K. H.; Ryu D. Y.; Choi J. Ecotoxicity of silver nanoparticles on the soil nematode Caenorhabditis elegans using functional ecotoxicogenomics. Environ. Sci. Technol. 2009, 43103933–40. [DOI] [PubMed] [Google Scholar]
- Scown T. M.; Santos E. M.; Johnston B. D.; Gaiser B.; Baalousha M.; Mitov S.; Lead J. R.; Stone V.; Fernandes T. F.; Jepson M.; van Aerle R.; Tyler C. R. Effects of aqueous exposure to silver nanoparticles of different sizes in rainbow trout. Toxicol. Sci. 2010, 1152521–34. [DOI] [PubMed] [Google Scholar]
- Choi J. E.; Kim S.; Ahn J. H.; Youn P.; Kang J. S.; Park K.; Yi J.; Ryu D. Y. Induction of oxidative stress and apoptosis by silver nanoparticles in the liver of adult zebrafish. Aquat. Toxicol. 2010, 1002151–9. [DOI] [PubMed] [Google Scholar]
- Nair P. M.; Choi J. Identification, characterization and expression profiles of Chironomus riparius glutathione S-transferase (GST) genes in response to cadmium and silver nanoparticles exposure. Aquat. Toxicol. 2011, 1013–4550–60. [DOI] [PubMed] [Google Scholar]
- Niazi J. H.; Sang B. I.; Kim Y. S.; Gu M. B. Global Gene Response in Saccharomyces cerevisiae Exposed to Silver Nanoparticles. Appl. Biochem. Biotechnol. 2011, 16481278–91. [DOI] [PubMed] [Google Scholar]
- Matsumura H.; Yoshida K.; Luo S.; Kimura E.; Fujibe T.; Albertyn Z.; Barrero R. A.; Kruger D. H.; Kahl G.; Schroth G. P.; Terauchi R. High-throughput SuperSAGE for digital gene expression analysis of multiple samples using next generation sequencing. PLoS ONE 2010, 58e12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull G. C.; Van Look K. J.; Santos E. M.; Filby A. L.; Gray D. M.; Nash J. P.; Tyler C. R. Variability in measures of reproductive success in laboratory-kept colonies of zebrafish and implications for studies addressing population-level effects of environmental chemicals. Aquat. Toxicol. 2008, 872115–26. [DOI] [PubMed] [Google Scholar]
- ISO, Water Quality—Determination of the Acute Lethal Toxicity of Substances to a Freshwater Fish [Brachydanio rerio Hamilton-Buchanan (Teleostei, Cyprinidae)], Part 3. Flow-Through Method; International Organization for Standardization, 1996. [Google Scholar]
- Osborne O. J.; Johnston B. D.; Moger J.; Balousha M.; Lead J. R.; Kudoh T.; Tyler C. R., Effects of particle size and coating on nanoscale Ag and TiO(2) exposure in zebrafish (Danio rerio) embryos. Nanotoxicology 2012, DOI: 10.3109/17435390.2012.737484. [DOI] [PubMed] [Google Scholar]
- Field D.; Tiwari B.; Booth T.; Houten S.; Swan D.; Bertrand N.; Thurston M. Open software for biologists: From famine to feast. Nat. Biotechnol. 2006, 247801–3. [DOI] [PubMed] [Google Scholar]
- Flicek P.; Amode M. R.; Barrell D.; Beal K.; Brent S.; Chen Y.; Clapham P.; Coates G.; Fairley S.; Fitzgerald S.; Gordon L.; Hendrix M.; Hourlier T.; Johnson N.; Kahari A.; Keefe D.; Keenan S.; Kinsella R.; Kokocinski F.; Kulesha E.; Larsson P.; Longden I.; McLaren W.; Overduin B.; Pritchard B.; Riat H. S.; Rios D.; Ritchie G. R.; Ruffier M.; Schuster M.; Sobral D.; Spudich G.; Tang Y. A.; Trevanion S.; Vandrovcova J.; Vilella A. J.; White S.; Wilder S. P.; Zadissa A.; Zamora J.; Aken B. L.; Birney E.; Cunningham F.; Dunham I.; Durbin R.; Fernandez-Suarez X. M.; Herrero J.; Hubbard T. J.; Parker A.; Proctor G.; Vogel J.; Searle S. M. Ensembl 2011. Nucleic Acids Res. 2011, 39Database issueD800–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B.; Trapnell C.; Pop M.; Salzberg S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 103R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson N.; Oveisi-Fordorei M.; Zuyderduyn S. D.; Varhol R. J.; Fjell C.; Marra M.; Jones S.; Siddiqui A. DiscoverySpace: An interactive data analysis application. Genome Biol. 2007, 81R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S.; Claverie J. M. The significance of digital gene expression profiles. Genome Res. 1997, 710986–95. [DOI] [PubMed] [Google Scholar]
- Storey J. D.; Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 2003, 100169440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W.; Sherman B. T.; Zheng X.; Yang J.; Imamichi T.; Stephens R.; Lempicki R. A., Extracting biological meaning from large gene lists with DAVID. Curr. Protoc. Bioinformatics 2009, Chapter 13, Unit 13 11. [DOI] [PubMed] [Google Scholar]
- Huang da W.; Sherman B. T.; Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4144–57. [DOI] [PubMed] [Google Scholar]
- Edgar R.; Domrachev M.; Lash A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 301207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD, OECD Guideline for the testing of chemicals. Draft proposal for a new guideline - Fish Embryo Toxicity (FET) Test, 2006. www.oecd.org.
- Vesterlund L.; Jiao H.; Unneberg P.; Hovatta O.; Kere J. The zebrafish transcriptome during early development. BMC Dev. Biol. 2011, 11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus Z.; Zakrzewska A.; Agoston V. C.; Ordas A.; Racz P.; Mink M.; Spaink H. P.; Meijer A. H. Deep sequencing of the zebrafish transcriptome response to mycobacterium infection. Mol. Immunol. 2009, 46152918–30. [DOI] [PubMed] [Google Scholar]
- Pan Q.; Shai O.; Lee L. J.; Frey B. J.; Blencowe B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40121413–5. [DOI] [PubMed] [Google Scholar]
- Wang E. T.; Sandberg R.; Luo S.; Khrebtukova I.; Zhang L.; Mayr C.; Kingsmore S. F.; Schroth G. P.; Burge C. B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 4567221470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. J.; Browning L. M.; Nallathamby P. D.; Desai T.; Cherukuri P. K.; Xu X. H. In vivo quantitative study of sized-dependent transport and toxicity of single silver nanoparticles using zebrafish embryos. Chem. Res. Toxicol. 2012, 2551029–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poynton H. C.; Lazorchak J. M.; Impellitteri C. A.; Blalock B. J.; Rogers K.; Allen J.; Loguinov A. V.; Heckman L.; Govindasmawy S. Toxicogenomic responses of nanotoxicity in Daphnia magna exposed to silver nitrate and coated silver nanoparticles. Environ. Sci. Technol. 2012, 46116288–96. [DOI] [PubMed] [Google Scholar]
- Nair P. M.; Choi J., Characterization of a ribosomal protein L15 cDNA from Chironomus riparius (Diptera; Chironomidae): Transcriptional regulation by cadmium and silver nanoparticles. Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol. 159, (3), 157-62. [DOI] [PubMed] [Google Scholar]
- Powers C. M.; Badireddy A. R.; Ryde I. T.; Seidler F. J.; Slotkin T. A. Silver nanoparticles compromise neurodevelopment in PC12 cells: Critical contributions of silver ion, particle size, coating, and composition. Environ. Health Perspect. 2011, 119137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt U. Energy converting NADH: Quinone oxidoreductase (complex I). Annu. Rev. Biochem. 2006, 75, 69–92. [DOI] [PubMed] [Google Scholar]
- Costa C. S.; Ronconi J. V.; Daufenbach J. F.; Goncalves C. L.; Rezin G. T.; Streck E. L.; Paula M. M. In vitro effects of silver nanoparticles on the mitochondrial respiratory chain. Mol. Cell. Biochem. 2010, 3421–251–6. [DOI] [PubMed] [Google Scholar]
- AshaRani P. V.; Low Kah Mun G.; Hande M. P.; Valiyaveettil S. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano 2009, 32279–90. [DOI] [PubMed] [Google Scholar]
- Hidalgo E.; Dominguez C. Study of cytotoxicity mechanisms of silver nitrate in human dermal fibroblasts. Toxicol. Lett. 1998, 983169–79. [DOI] [PubMed] [Google Scholar]
- Teodoro J. S.; Simoes A. M.; Duarte F. V.; Rolo A. P.; Murdoch R. C.; Hussain S. M.; Palmeira C. M. Assessment of the toxicity of silver nanoparticles in vitro: A mitochondrial perspective. Toxicol. In Vitro 2011, 253664–70. [DOI] [PubMed] [Google Scholar]
- Anastasiou D.; Poulogiannis G.; Asara J. M.; Boxer M. B.; Jiang J. K.; Shen M.; Bellinger G.; Sasaki A. T.; Locasale J. W.; Auld D. S.; Thomas C. J.; Vander Heiden M. G.; Cantley L. C. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 2011, 33460601278–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka R. B.; Chandel N. S. Warburg Effect and Redox Balance. Science 2011, 33460601219–1220. [DOI] [PubMed] [Google Scholar]
- Yang X.; Gondikas A. P.; Marinakos S. M.; Auffan M.; Liu J.; Hsu-Kim H.; Meyer J. N. Mechanism of silver nanoparticle toxicity is dependent on dissolved silver and surface coating in Caenorhabditis elegans. Environ. Sci. Technol. 2012, 4621119–27. [DOI] [PubMed] [Google Scholar]
- Agarwal D. P.; Goedde H. W. Pharmacogenetics of alcohol dehydrogenase (ADH). Pharmacol. Ther. 1990, 45169–83. [DOI] [PubMed] [Google Scholar]
- Reimers M. J.; Hahn M. E.; Tanguay R. L. Two zebrafish alcohol dehydrogenases share common ancestry with mammalian class I, II, IV, and V alcohol dehydrogenase genes but have distinct functional characteristics. J. Biol. Chem. 2004, 2793738303–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasmahapatra A. K.; Doucet H. L.; Bhattacharyya C.; Carvan M. J. 3rd Developmental expression of alcohol dehydrogenase (ADH3) in zebrafish (Danio rerio). Biochem. Biophys. Res. Commun. 2001, 28651082–6. [DOI] [PubMed] [Google Scholar]
- Tamai T. K.; Young L. C.; Whitmore D. Light signaling to the zebrafish circadian clock by Cryptochrome 1a. Proc. Natl. Acad. Sci. U. S. A. 2007, 1043714712–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.