Table 1. Interaction ranks distribution for the Mintseris Benchmark 2.0.
| Mintseris Benchmark 2.0 - 168 proteins | |||||
| Top % | # top proteins | 168 vs 168 (%) | 56 vs 168 (%) | ||
| exp | pred | exp | pred | ||
| 1 | 1 | 42 (25) | 6 (4) | 16 (29) | 3 (5) |
| 5 | 8 | 76 (45) | 23 (14) | 28 (50) | 9 (16) |
| 10 | 17 | 98 (58) | 41 (25) | 36 (64) | 17 (31) |
| 15 | 25 | 118 (70) | 50 (30) | 45 (80) | 20 (36) |
| 20 | 34 | 126 (75) | 59 (36) | 45 (80) | 23 (42) |
| 30 | 50 | 136 (81) | 76 (46) | 47 (84) | 33 (60) |
| 40 | 67 | 145 (86) | 98 (59) | 49 (88) | 38 (69) |
| 50 | 84 | 154 (92) | 117 (70) | 54 (94) | 41 (75) |
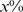 of complexes obtained by docking the proteins with all 168 proteins in the environment (this means that the NII score of the native complex falls in the top
of complexes obtained by docking the proteins with all 168 proteins in the environment (this means that the NII score of the native complex falls in the top 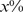 scores). Native complexes identification is realized either by knowing the experimental interface (exp) or by predicting it (pred). Cumulative counts and percentages are displayed. The selected set of 56 monomers considered in [33] is also evaluated against the 168 proteins (fifth and sixth columns). The number of top proteins corresponding to the
scores). Native complexes identification is realized either by knowing the experimental interface (exp) or by predicting it (pred). Cumulative counts and percentages are displayed. The selected set of 56 monomers considered in [33] is also evaluated against the 168 proteins (fifth and sixth columns). The number of top proteins corresponding to the 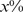 of the total number of proteins in the specified environment is given (second column). Over the 168 proteins in the Mintseris dataset, we report the number of proteins (third and fourth columns) whose native complex is identified within the top
of the total number of proteins in the specified environment is given (second column). Over the 168 proteins in the Mintseris dataset, we report the number of proteins (third and fourth columns) whose native complex is identified within the top
