Table 2. Partner prediction based on the exploration of the full conformational space.
| Protein dataset | Experimental interfaces | Predicted interfaces | |||||||
| Subset type | # proteins | JET+NIP | NIP | JET | |||||
| AUC | Sen | Spe | AUC | Sen | Spe | AUC | AUC | ||
| Mintseris DB | 168 | 0.84 | 52 | 92 | 0.61* | 25* | 89* | 0.53* | 0.59* |
| Enzyme-Inhibitor & Others | 124 | 0.84 | 54 | 92 | 0.66* | 34* | 87* | 0.56* | 0.65* |
| Enzyme-Inhibitor | 46 | 0.85 | 59 | 88 | 0.77 | 65 | 78 | 0.60 | 0.72 |
| Antibody-Antigen | 20 | 0.89 | 95 | 66 | 0.58 | 15 | 70 | 0.61 | 0.52 |
| Antigen-Bound Antibody | 24 | 0.91 | 79 | 80 | 0.56 | 38 | 74 | 0.63 | 0.53 |
| Others | 78 | 0.84 | 62 | 89 | 0.61* | 25* | 88* | 0.52* | 0.62* |
| Rigid | 126 | 0.87 | 59 | 91 | 0.60* | 29* | 85* | 0.53* | 0.59* |
| Medium | 26 | 0.85 | 73 | 81 | 0.68 | 58 | 80 | 0.53 | 0.67 |
| Difficult | 16 | 0.77 | 69 | 78 | 0.65 | 38 | 80 | 0.66 | 0.63 |
| Monomeric (both partners) | 86 | 0.87 | 66 | 89 | 0.63 | 36 | 85 | 0.55 | 0.63 |
| Multimeric (at least one partner) | 82 | 0.81 | 59 | 88 | 0.61* | 32* | 86* | 0.51* | 0.61* |
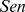 ) and specificity (
) and specificity (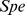 ) are also given at a NII threshold cutoff of 0.5 for predictions based on experimental interfaces, and at a NII threshold cutoff of 0.25 for predicted interfaces. Calculations based on JET and NIP predicted interfaces use weights
) are also given at a NII threshold cutoff of 0.5 for predictions based on experimental interfaces, and at a NII threshold cutoff of 0.25 for predicted interfaces. Calculations based on JET and NIP predicted interfaces use weights 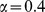 ,
, 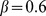 (see Methods). The * symbol refers to values computed on subsets that have been cleaned of the complex 1ML0 for which JET provided no interaction site (leading to a
(see Methods). The * symbol refers to values computed on subsets that have been cleaned of the complex 1ML0 for which JET provided no interaction site (leading to a 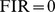 because of no common residue between the small predicted interface and the docked one). The Mintseris dataset and the subsets Enzyme-Inhibitor & Others, Others, Rigid and Multimeric contain 166, 122, 76, 124 and 80 proteins, respectively. See also Tables S8–S9 in Text S1 for other threshold cutoffs and performance measures. The analysis is realized by assuming knowledge of either the experimental interfaces or the predicted interfaces. Performance of partner prediction is evaluated through AUC values computed on the Mintseris dataset and its different subsets. Sensitivity (
because of no common residue between the small predicted interface and the docked one). The Mintseris dataset and the subsets Enzyme-Inhibitor & Others, Others, Rigid and Multimeric contain 166, 122, 76, 124 and 80 proteins, respectively. See also Tables S8–S9 in Text S1 for other threshold cutoffs and performance measures. The analysis is realized by assuming knowledge of either the experimental interfaces or the predicted interfaces. Performance of partner prediction is evaluated through AUC values computed on the Mintseris dataset and its different subsets. Sensitivity (
