Table 3. Interaction ranks distribution for the Mintseris' Enzyme-Inhibitors dataset.
| MAXDo vs HEX - Enzyme-Inhibitors dataset | |||||
| Top % | # top proteins | MAXDo | HEX | ||
| exp | pred | exp | pred | ||
| 1 | 1 | 9 (20) | 4 (9) | 7 (15) | 2 (4) |
| 5 | 2 | 16 (35) | 10 (22) | 16 (35) | 5 (11) |
| 10 | 5 | 24 (52) | 21 (46) | 27 (59) | 6 (13) |
| 15 | 7 | 27 (59) | 25 (54) | 30 (65) | 8 (17) |
| 20 | 9 | 28 (61) | 28 (61) | 35 (76) | 8 (17) |
| 30 | 14 | 35 (76) | 33 (72) | 37 (80) | 18 (39) |
| 40 | 18 | 38 (83) | 36 (78) | 38 (83) | 28 (61) |
| 50 | 23 | 41 (89) | 41 (89) | 41 (89) | 33 (72) |
+JET scores obtained with weights 0.4 and 0.6 for NIP and JET respectively. For each CC-D, over the 46 Enzyme-Inhibitors in the Mintseris dataset, we report the number of proteins whose native complex is identified within the top 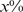 of complexes obtained by docking the protein with all 46 proteins in the environment. Cumulative counts and percentages (in parenthesis) are displayed. See legend of Table 1. CC-D has been realized with MAXDo and HEX v6.3 docking algorithms. Calculations based on predicted interfaces (fourth and sixth columns) are made with NIP
of complexes obtained by docking the protein with all 46 proteins in the environment. Cumulative counts and percentages (in parenthesis) are displayed. See legend of Table 1. CC-D has been realized with MAXDo and HEX v6.3 docking algorithms. Calculations based on predicted interfaces (fourth and sixth columns) are made with NIP
