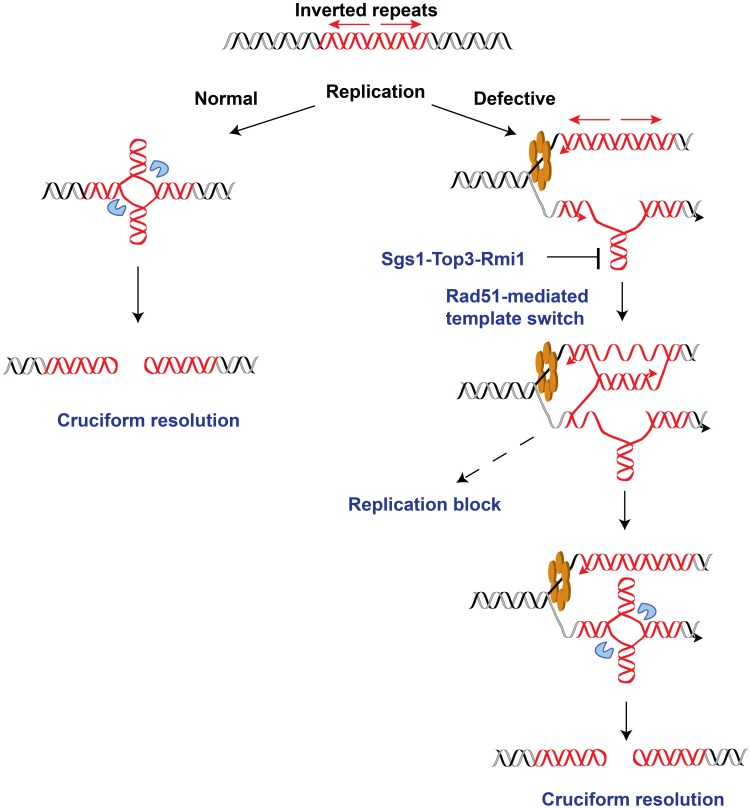
Figure 6. Model for Alu-IRs-mediated fragility under conditions of replication proficiency and deficiency.
The red helixes, blue pacman and orange hexamer depict the inverted repeats, the putative nuclease and the DNA replication helicase, respectively. In the case of normal replication, cruciform structure might form outside of S-phase as a result of chromatin packing or remodeling. On the other hand, long single-stranded DNA exposed due to compromised replication would facilitate the formation of a hairpin, which could further be converted into cruciform structure via template switch by Rad51. The intermediates of template switching present a strong obstacle for the replication machinery that is manifested as replication block in the replication-deficient strains. The Sgs1-Top3-Rmi1 dissolvosome might participate in unwinding the hairpin. Once formed, the cruciform structure might be attacked by the putative nuclease, leading to DSBs at the IRs.