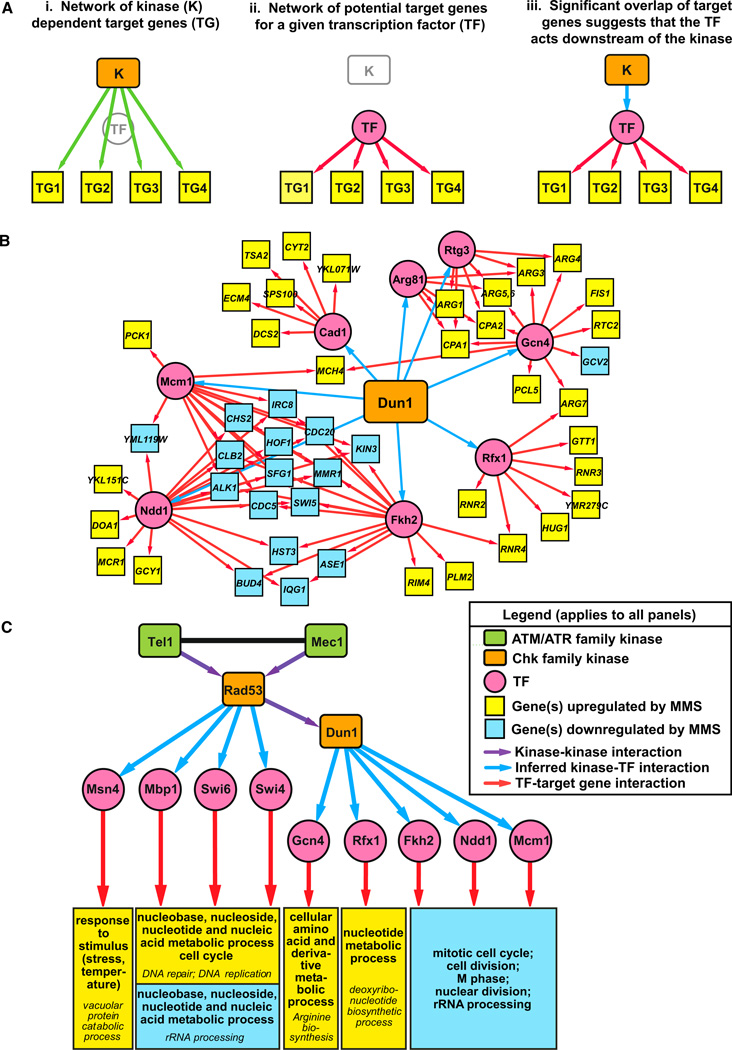
Figure 3. Implicating TFs as Downstream Effectors of the Checkpoint-Kinase-Dependent Transcriptional Response to DNA Damage.
(A) Strategy used to identify kinase-TF interactions. (i) The kinase transcriptional regulatory network consists of interactions between the kinase and the set of target genes with kinase-dependent changes in gene expression. (ii) The TF regulatory network consists of predicted interactions between TFs and their target genes (Beyer et al., 2006). (iii) Significant overlap of target genes in the kinase and TF regulatory networks suggests that an interaction between the kinase and TF explains how the kinase regulates transcription of its target genes.
(B) The transcriptional regulatory network generated by applying this analysis to the Dun1-dependent set of target genes.
(C) A global transcriptional regulatory network showing kinase-TF interactions mediating the checkpoint-kinase-dependent transcriptional response to DNA damage. Kinase-TF interactions were only included in this network if four of the seven checkpoint-kinase-dependent gene sets showed enrichment for targets of the TF (see Table S6 for enrichment analysis and Figure S1 for a larger network that includes TFs enriched in two or more kinase-dependent gene sets). For simplicity, the target genes were replaced with modules showing the enriched GO terms in the sets of Rad53-dependent genes predicted to be targets of the TF (selected from Table S7). Fkh2, Ndd1, and Mcm1 form a transcriptional complex, and Swi6 forms distinct transcriptional complexes with Mbp1 (MBF) and Swi4 (SBF). Therefore, the Fhh2/Ndd1/Mcm1 and MBF/SBF target genes are represented in overlapping modules of enriched GO terms (see Figure S2). Fkh1 was omitted because nearly all of its predicted targets comprise a subset of the target genes of the Fkh2/Ndd1/Mcm1 complex. GO terms in italics are subcategories of the main GO terms listed above.
See also Tables S6 and S7 and Figures S1 and S2.