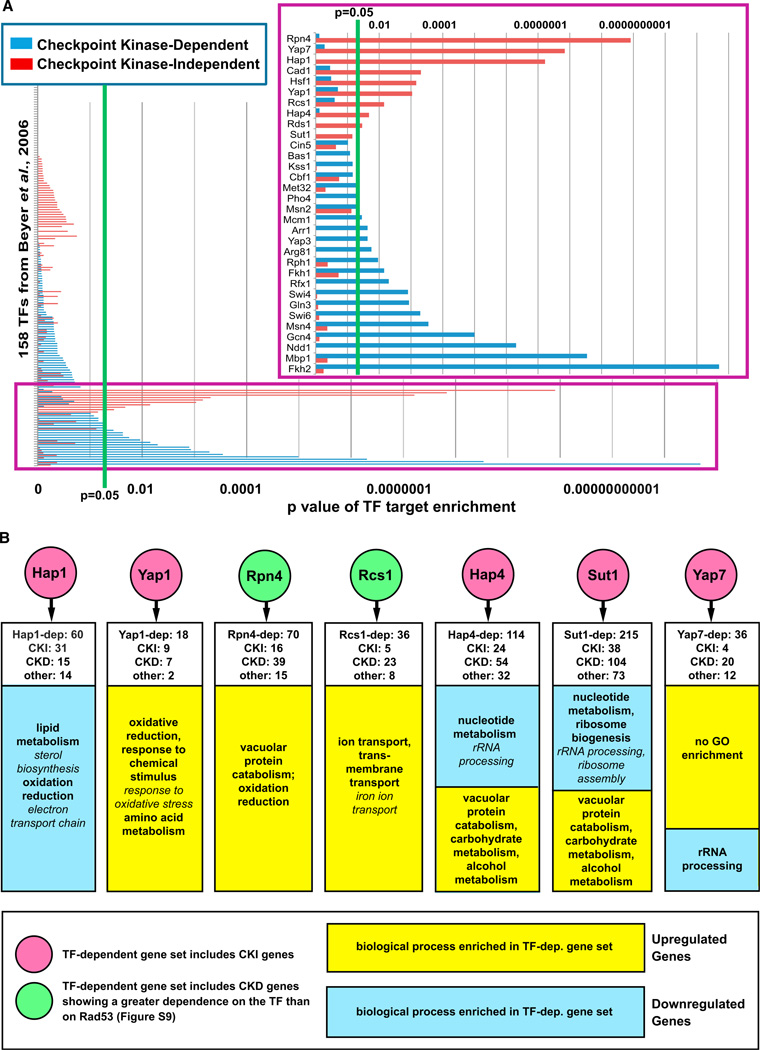
Figure 6. Identification of TFs Mediating Checkpoint-Kinase-Independent Gene Expression in Response to MMS Treatment.
(A) The set of genes that was checkpoint kinase dependent (i.e., showed reduced differential expression in at least two of the checkpoint kinase mutants) and the set of genes that was checkpoint-kinase- independent (i.e., not affected by any of the checkpoint kinase mutations) show enrichment for targets of distinct sets of TFs. Enrichment for TF targets was performed as in Figure 3, and the p values for enrichment for targets of all 158 TFs are shown here and in Table S10. The inset shows TFs with enrichment p values that are <0.1.
(B) Expression profiling confirms that the predicted TFs regulate checkpoint-kinase-independent (CKI) gene expression and identifies checkpoint-kinase-dependent (CKD) genes regulated by the TFs (see Table S2 and Figures S8–S10 for expression profile data). TF-dependent genes for Hap1, Hap4, Rcs1, Sut1, and Yap7 were identified by expression profiling of TF mutants as in Figure 2 (see Table S3), whereas deletion-buffered genes from Workman et al. (2006) were analyzed for Rpn4 and Yap1. White boxes indicate the number of CKD and CKI genes that showed reduced differential expression in the TF mutant. GO terms in italics are subcategories of the main GO terms listed.