Abstract
The bifunctional enzyme thymidylate synthase–dihydrofolate reductase (TS–DHFR) from the protozoal parasite Cryptosporidium hominis is a potential molecular target for the design of antiparasitic therapies for AIDS-related opportunistic infections. The enzyme exists as a homodimer with each monomer containing a unique swap domain known as a “crossover helix” that binds in a cleft on the adjacent DHFR active site. This crossover helix is absent in species containing monofunctional forms of DHFR such as human. An in-depth understanding of protein–protein interactions between the crossover helix and adjacent DHFR active site that might modulate enzyme integrity or function would allow for insights into rational design of species-specific allosteric inhibitors. Mutational analysis coupled with structural studies and biophysical and kinetic characterization of crossover helix mutants identifies this domain as essential for full enzyme stability and catalytic activity, and pinpoints these effects to distinct faces of the crossover helix important in protein–protein interactions. Moreover, targeting this helical protein interaction with α-helix mimetics of the crossover helix leads to selective inhibition and destabilization of the C. hominis TS–DHFR enzyme, thus validating this region as a new avenue to explore for species-specific inhibitor design.
Introduction
Dihydrofolate reductase (DHFR) is a well-studied enzyme that catalyzes the reduction of dihydrofolate (H2folate) to tetrahydrofolate (H4folate) utilizing the reducing cofactor NADPH. There is a wealth of information available on the structure, dynamics, and inhibition of DHFR from many species. However, the DHFR domain from the protozoan enzyme thymidylate synthase–dihydrofolate reductase (TS–DHFR)‡ is structurally unique. Unlike the monofunctional DHFR from most species such as humans and E. coli, TS–DHFR from protozoan parasites is bifunctional, encoding both TS and DHFR domains on the same polypeptide connected by a linker of varying length.1 Several of these obligate, intracellular protozoal parasites are members of the Apicomplexa phylum and include Plasmodium falciparum, Toxoplasma gondii, and Cryptosporidium hominis. While P. falciparum is the causative agent of malaria, T. gondii and C. hominis are parasites responsible for AIDS-related opportunistic infections.
The bifunctional enzymes from these apicomplexans all have long TS-to-DHFR linkers. The three dimensional structures are available for the P. falciparum and C. hominis enzymes and these structures reveal that contained within the linker is a unique domain termed a “crossover helix”, which packs against the backside of the DHFR active site.2,3 Essentially a swap domain, the crossover helix is involved in protein–protein interactions between one DHFR monomer and the active site of the adjacent monomer. While kinetic studies on the bifunctional enzyme from the malaria parasite P. falciparum showed this domain to have no effect on the enzyme catalysis,4 previous work from our lab indicates that the TS–DHFR crossover helix in C. hominis, is necessary for full DHFR activity and results in a dramatic impairment in catalytic rate when mutated.5
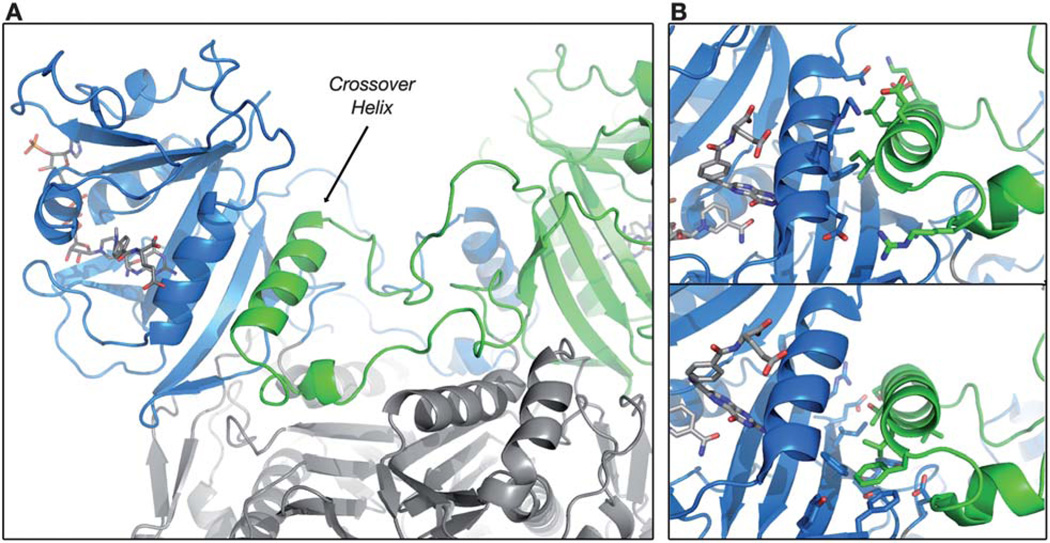
The position of the crossover helix in C. hominis TS–DHFR is detailed in Fig. 1A. Whereas the crossover helix in P. falciparum TS–DHFR makes few contacts with the DHFR domain, the C. hominis TS-to-DHFR linker inserts the crossover helix into a cleft at the backside of the adjacent DHFR active site. The crossover helix makes extensive interactions both with Helix B of the DHFR active site and the beta sheet domain more distal to the active site (Fig. 1B). Interestingly, two orthogonal faces of the helix contact these two subdomains separately. While the structure highlights the intimate contacts between nominal DHFR and this swap domain, little is known about potential mechanisms by which the domain may modulate enzyme function.
Fig. 1.
Crystal structure of the C. hominis DHFR domain highlighting the position of the crossover helix. (A) The orientation of the two DHFR domains (blue and green) and the TS domain (gray) of C. hominis TS–DHFR are illustrated in cartoon form (from PDB ID 2OIP). The crossover helix (green) is shown packing against the opposite DHFR domain (blue), between Helix B of the active site and the beta sheet domain. DHFR ligands are shown as gray sticks. (B) Close-up view of specific crossover helix interactions. Top panel, the Helix Face of the crossover helix (residues as green sticks: K194, D198, L202, E205, I206, R210) interacts with residues of Helix B. Bottom panel, the Beta Face of the crossover helix (residues: S195, I196, T199, V200, L203, F207) interacts with residues of the beta sheet domain. Residues interacting with the faces are shown as blue sticks.
Cryptosporidium hominis causes a serious gastrointestinal disease in AIDS patients and other immunocompromised individuals, is especially dangerous against children and the elderly, and has recently been classified as a category B biodefense pathogen,6,7 however there are few effective treatments for the parasite.8,9 Although DHFR is an essential enzyme and is a validated drug target in other parasitic protozoa, the currently available therapeutics have proven to be ineffective against C. hominis DHFR. With the exception of a recent study,10 efforts to increase potency have often led to a decrease in species-specificity.11,12 The idea that a novel allosteric site important for catalysis may be targeted for species-specific inhibitor design is an attractive prospect. In a previous validation study, we utilized virtual screening to target a pocket near the crossover helix and were able to find micromolar-range inhibitors of Cryptosporidium hominis thymidylate synthase–dihydrofolate reductase, ChTS–DHFR.13 A more detailed, comprehensive knowledge of the role of the crossover helix in enzyme function, and data on the specific interactions important for this effect, are critical in guiding the initial stages of a rational search for novel allosteric inhibitors of ChTS–DHFR.
In this study, we utilized a steady-state and transient kinetic approach to characterize mutants of the crossover helix that may be involved in key protein–protein interactions and identify the individual faces of the helix that modulate catalytic activity and stability of the TS–DHFR enzyme. These studies guided rationally designed benzoylurea α-helix mimetics that allosterically inhibit the enzyme and compromise overall enzyme stability. The results presented here provide in-depth insights into the role of this critical crossover helix in a novel bifunctional DHFR, define a molecular mechanism for modulating enzyme catalysis, and highlight the potential to utilize biochemical information of a crucial protein–protein interaction to rationally design an enzyme inhibitor.
Results
Validation of the crossover helix as essential for ChTS–DHFR activity and stability
Alanine mutations were made to the crossover helix on two faces of the helix; the 6 amino acid residues comprising the side facing Helix B of the DHFR active site (denoted Helix Face mutant), and the 6 residues comprising the side facing the beta-sheet of DHFR (denoted Beta Face mutant) (Fig. 1B). A third all alanine crossover helix mutant enzyme was also examined. Alanine mutagenesis was chosen in order to specifically disrupt amino acid interactions, while still maintaining helical character.14 The residues comprising the crossover helix were predicted by nnPredict to be helical for all mutants.15
Recent work from this lab showed that both the Helix Face mutant and the All Alanine mutant enzymes display have impaired catalytic activity.13 It was unclear whether interactions on the orthogonal beta-sheet face of the helix were important for enzyme activity. Fig. 2A shows the complete steady-state analysis for crossover helix mutant enzymes. Mutations of either face of the helix or the entire face of the helix all cause a roughly 2-fold decrease in DHFR activity. Interestingly, all of the mutants also cause an unexpected decrease in activity of the TS domain, the active site of which resides 30 Å away from the crossover helix (Fig. 2B).
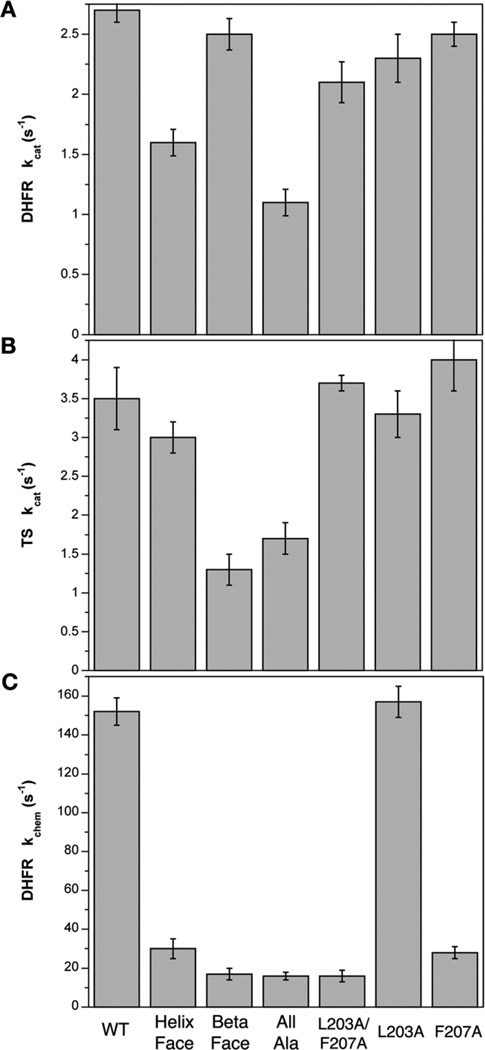
Fig. 2.
Impaired rates of catalysis in the crossover helix mutant enzymes. (A) DHFR steady-state turnover rates (kcat) determined by a spectroscopic assay (WT = 2.7 s−1; Helix Face = 1.6 s−1; Beta Face = 2.5 s−1; All Alanine = 1.1 s−1; L203A/F207A = 2.1 s−1; L203A = 2.3 s−1; F207A = 2.5 s−1). Values are all represented as the average ± standard deviation. (B) TS steady-state turnover rates (WT = 3.5 s−1; Helix Face = 3.0 s−1; Beta Face = 1.3 s−1; All Alanine = 1.7 s−1; L203A/F207A = 3.7 s−1; L203A = 3.3 s−1; F207A = 4.0 s−1). (C) Single turnover experiments monitoring the rate of hydride transfer, or kchem, conducted utilizing stopped-flow fluorescence (WT = 152 s−1; Helix Face = 30 s−1; Beta Face = 17 s−1; All Alanine = 16 s−1; L203A/F207A = 16 s−1; L203A = 157 s−1; F207A = 28 s−1).
In DHFR catalysis, the rate-limiting step is product release, and therefore measuring steady-state activity does not allow the precise measurement of the chemistry step, hydride transfer. In order to assess the importance of the crossover helix on the rate of chemistry, a stopped-flow fluorescence approach was utilized. The DHFR co-factor NADPH and an active site tryptophan form a FRET pair that is lost upon NADPH consumption. Therefore, on a millisecond time scale, we can monitor DHFR chemistry corresponding to a loss in fluorescent signal (Fig. S1†). Single-enzyme-turnover experiments were conducted using this approach, yielding the rate of hydride transfer for WT (150 ± 7 s−1) and for each mutant (Fig. 2C). Interestingly, both the Helix Face and Beta Face mutant enzymes displayed a similar kinetic defect as the All Alanine enzyme (16 ± 2 s−1). This implies that both faces of the crossover helix, the face interacting with the active site and the face interacting with regions distal to the active site, are important for enzyme function.
An important feature we noted early on with the crossover helix mutant enzymes in comparison with WT enzyme was the relatively low yield of protein during expression and purification. The yield of the mutants was between 0.02 and 0.5 mg per liter of growth for the mutant enzymes, compared with ~4 mg L−1 for WT. Growth at lower temperatures increased the overall yield of the mutants, signifying a defect in thermal stability for these mutants. To assess the overall stability of the enzymes, we conducted thermal denaturation of the proteins, monitored by circular diochroism (CD) absorption at 222 nm. Fig. 3A shows the decrease in the fraction of folded enzyme as a function of temperature. All of the mutants showed a significant decrease in thermal stability compared to WT. Using a single-step transition model to estimate the melting temperature, the mutants showed an average decrease in stability of 12 °C compared to WT. Importantly, at 37 °C, all of the mutant enzymes were partially unfolded, whereas the WT enzyme was fully folded.
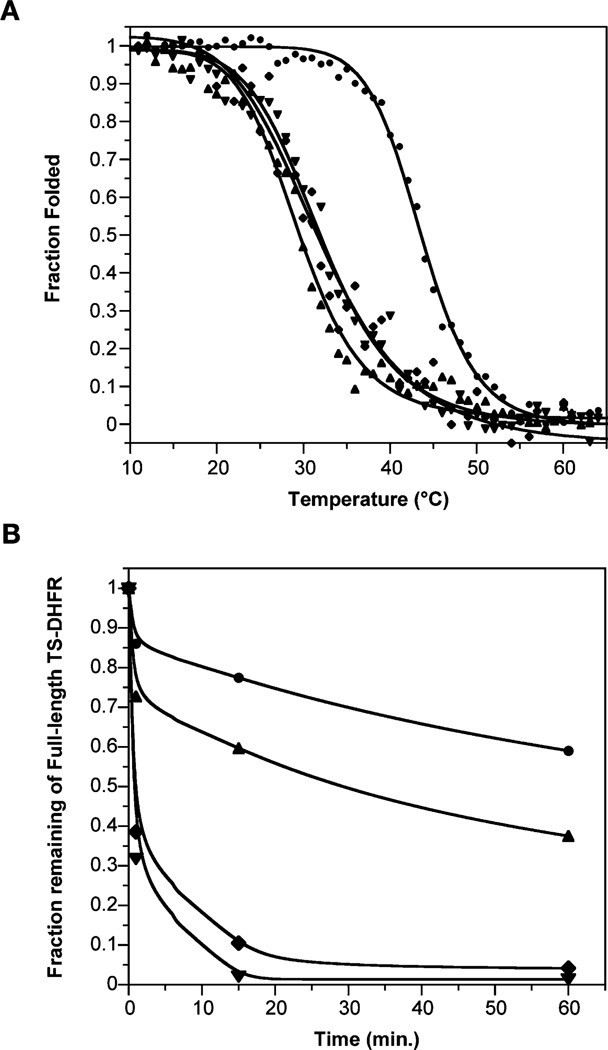
Fig. 3.
Decreased stability of the crossover helix mutant enzymes. (A) Thermal denaturation monitored by circular dichroism spectroscopy shows the melting transitions of WT TS–DHFR (●, Tm = 43.6 °C), Helix Face (▲, 29.5 °C), Beta Face (▼, 32.0 °C), and All Alanine (◆, 31.9 °C). Data were fit to a single transition curve. (B) Quantitation of limited trypsin proteolysis of WT and mutant enzymes. Values are normalized to starting full-length enzyme at time zero. The symbols are as in (A).
Using size-exclusion chromatography we saw no effect of this stability decrease on dimer formation to a lower detection limit of 200 nM (data not shown). However, as protein instability can also correlate with protease susceptibility,16,17 and this has been specifically demonstrated for monofunctional TS,18 we sought to determine the degree to which the mutant enzymes might be cleaved by common proteases. Limited proteolysis was conducted with trypsin, in which partial digestion of proteins was quenched at different time points and run on SDS-PAGE. Interestingly, all of the mutant enzymes were significantly more sensitive than WT to trypsin proteolysis (Fig. S2†). The experiments were repeated for Glu-C protease and showed a similar trend, indicating that the sensitivity is not protease-dependent. The relative amount of proteolysis for each protein was quantitated by measuring the decrease in intensity of the full-length band as a function of time (Fig. 3B). Overall, the mutants showed dramatic increases in trypsin susceptibility. The All Alanine and Beta Face mutants showed virtually no full-length protein left at 15 minutes, while WT retained nearly 80% of full length protein. Interestingly, the Helix Face enzyme showed only moderate sensitivity compared with the other mutants, retaining nearly 60% of full-length protein at 15 minutes.
Identification of F207A as critical for DHFR chemistry
The initial mutants were part of a larger-scale mutagenesis in which entire faces of the helix were mutated with the goal of disrupting entire interaction surfaces. To more specifically focus on the underlying mechanism by which the crossover helix may support catalytic function, we sought to pinpoint integral amino acids that may disrupt catalytic activity. Accordingly, we examined mutants of this region containing individual or a small number of amino acid changes. Our analysis of several enzymes containing a smaller number of mutations showed that no individual stretch of residues appeared to have a major effect on steady-state rate, as they each appeared to display only small decreases in kcat (data not shown). However, we were able to narrow down the defect in hydride transfer to the double mutant L203A/F207A, and further to a single residue, F207A, that displays nearly the entire decrease in catalytic activity as the helix mutants containing a larger number of mutations, at 28 ± 5 s−1 (Fig. 2C). Importantly, there was only minimal decrease in steady-state rate, implying that the effect of this residue is specific to the rate of hydride transfer. The stability of the F207A mutant enzyme was characterized as with the earlier crossover helix mutants, and did show a decrease in stability of 5 °C compared to WT, although showed no significant increase in protease sensitivity (Fig. S3†).
Phe207 resides at the base of the crossover helix, and interacts primarily with two phenylalanines in the beta-sheet domain of DHFR: Phe163 and Phe172. To determine if Phe207 communicates with the active site by direct modulation of these residues, the two corresponding alanine mutant enzymes (F163A and F172A) were expressed and analyzed. Unlike F207A, both the steady-state rates (2.5 s−1 and 2.6 s−1, respectively) and the pre-steady-state hydride transfer rates (169 s−1 and 154 s−1, respectively) were not significantly different than WT.
Toward elucidating a structural basis for the loss in activity of the crossover helix mutant enzymes, we have conducted crystallization trials with all of the mutants discussed. Likely due to thermal and/or conformational instability, the enzymes containing a larger number of mutations did not appear to crystallize under a varying range of conditions. However, we were able to grow 50 µm crystals of F207A mutant TS–DHFR enzyme in complex with DHFR and TS ligands in only one week, and determine the structure at 2.7 Å. Pertinent crystallographic values are included in Table 1.
Table 1.
Data collection and refinement statistics for the structure of ChTS–DHFR F207A mutant enzyme complexed with dUMP, CB3717, NADPH, and methotrexate
| Values in parentheses are for highest resolution shell | |
|---|---|
| Resolution limit (Å) | 2.70 |
| Space group | P3221 |
| Unit cell parameters (Å, °) | 121.9, 121.9, 342.3, 90, 90, 120 |
| No. reflections used | 77 069 |
| Completeness (%) | 99.4 (95.1) |
| Redundancy | 5.4 (5.4) |
| I/sig I | 13.8 (3.6) |
| Rmerge (%) | 8.1 (49.1) |
| No. monomers in asymmetric unit | 4 |
| Refinement statistics | |
| Rfactor (%) | 22.2 |
| Rfree (%) | 27.3 |
| Total no. of atoms | 17 282 |
| No. water molecules | 297 |
| RMSD, bonds (Å) | 0.009 |
| RMSD, angles (°) | 1.4 |
| Wilson B factor (Å2) | 55 |
| Coordinate error, Luzzati (Å) | 0.36 |
| Ramachandran plot statistics | |
| Residues in most favored regions (%) | 88.3 |
| Residues in additionally allowed (%) | 10.9 |
| Residues in generously allowed (%) | 0.7 |
| Residues in disallowed regions (%) | 0.1 |
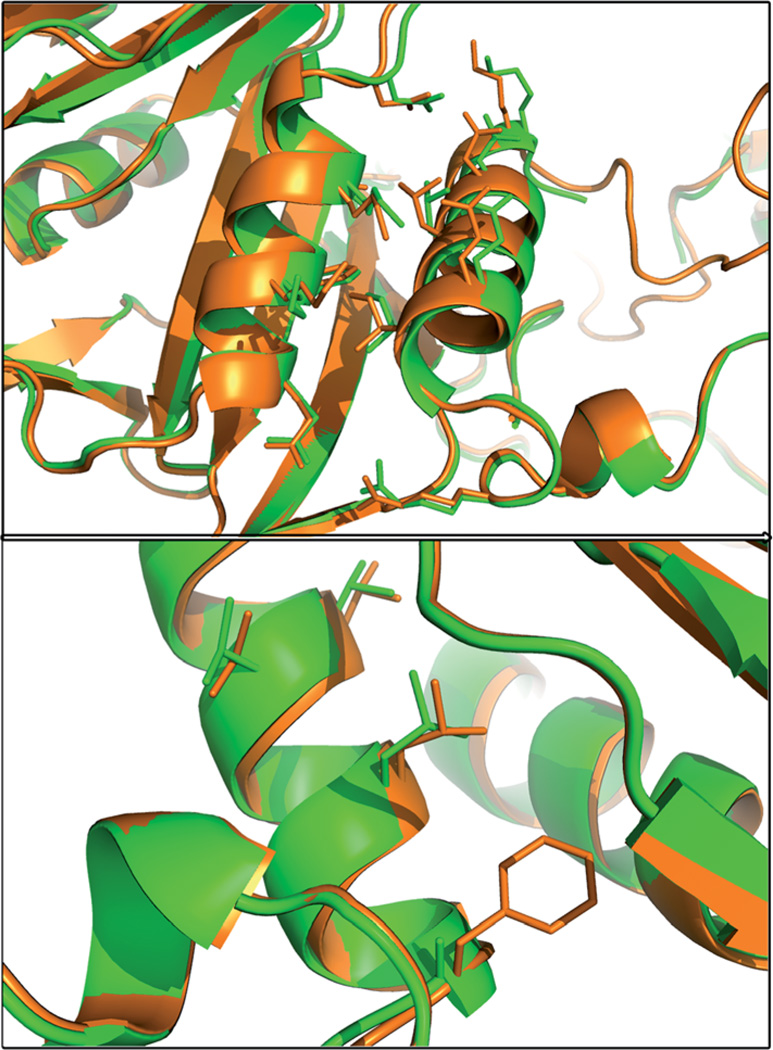
Compared with the WT crystal structure, there were no global conformational changes in the F207A mutant enzyme at either the DHFR or TS domains. Overall, the tertiary and secondary structure motifs were maintained. However, there was a small but significant change in the position of the crossover helix and Helix B. The crossover helix is shifted up and away from Helix B of the DHFR active site, and Helix B is shifted slightly away from the active site, though only at its C-terminus. The effects on the interacting amino acid residues of the crossover helix are shown in Fig. 4. All of the residues on the Helix Face are translated away from the DHFR domain, by an average change in the position of the alpha-carbon of 1.5 Å. The interacting residues near the C-terminus of Helix B are also shifted, but by less than 0.8 Å. The residues near the N-terminus of Helix B are significantly altered, with the side chains moved away from the crossover helix. Additionally, the residues on the beta-sheet face of the crossover helix are shifted by an average of 1 Å, although the positions of the interacting residues on the beta-sheet do not appear to be changed. From a structural perspective, the end result is an overall decrease in the extent of protein–protein interactions of the crossover helix with Helix B and the beta sheet. Therefore, it appears that Phe207 acts as an anchor to mediate the helix–helix interactions important for catalysis.
Fig. 4.
Altered crossover helix positioning in the crystal structure of the F207A mutant a least-squares alignment of F207A (green) with WT ChDHFR (orange) shows a slight but significant shift in the position of the helix. The alterations in the position of residues of the Helix Face (top panel) and Beta Face (bottom panel) are highlighted.
Selection and identification of crossover helix-mimetics as inhibitors of ChTS–DHFR
The analysis presented thus far demonstrates that the crossover helix region is important for full enzyme activity and overall enzyme stability. Since the crossover helix binds in a pocket distal to the DHFR active site, the area is intriguing as a novel site for inhibitors. Our data points to the exciting possibility that an inhibitor designed toward this region may also decrease enzyme stability. Importantly, our mutational data can inform our choices of potential small molecule inhibitors.
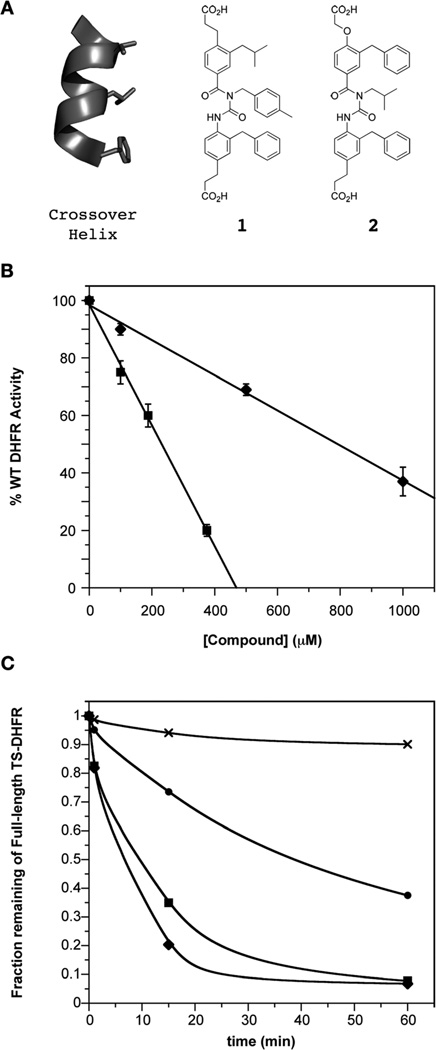
One attractive idea for designing inhibitors of this region involves using a synthetic mimic of the crossover helix to bind in the non-active site cleft and thus compete with the crossover helix-to-DHFR interaction. Recent work on α-helix mimetics has involved the use of benzoylurea oligomers, which utilize intramolecular hydrogen bonds to favor a linear conformation and allow projection of side chains analogous to that of a natural peptide helix (Fig. 5A). This approach has proven successful for inhibiting the Bcl-xL/Bak protein–protein interaction.19
Fig. 5.
Benzoylurea α-helix mimetics as novel inhibitors of ChTS–DHFR. (A) Crossover helix is shown in gray, with the side chain residues to mimic shown as sticks (V200, L203, F207). Benzyolurea mimetics 1 (LxxFxxxF) and 2 (FxxLxxxF) contain a hydrophobic face and correct phenylalanine side-chain projection for position 207. (B) Dose–response curves of mimetics 1 (◆) and 2 (■), plotted as percent of WT ChDHFR activity remaining. Data fit to linear equation. (C) Quantitation of limited trypsin proteolysis of ChTS–DHFR in the absence (●) and presence of 2000 µM mimetic 1 (◆), 750 µM mimetic 2 (■), and 100 µM methotrexate (×). Based on binding affinities, the concentrations are near-saturating for the helix mimetics, and saturating for methotrexate. Line is a smooth fit.
In theory, any sequence of i, i + 3/4, and i + 7 residues in an α-helix could serve as a basis for an α-helix mimetic. Therefore we could use as a model amino acids on either face of the crossover helix as shown in Fig. 1B. However, our mutational data indicate that the Helix Face mediates steady-state activity, so it is that interaction that we targeted for disruption of catalytic activity. Additionally, the thermal stability data and crystal structure presented establish that Phe207 is critical for proper alignment of the entire crossover helix into its binding cleft, and therefore would likely aid in positioning an α-helix mimetic. Therefore, we used as a template the stretch of residues on the beta-sheet face (V200xxL203xxxF207) to rationally design helix mimics. We hypothesized a possible mechanism of inhibition by which the helix mimetic would compete with the crossover helix for the beta-sheet interactions, displace the crossover helix in doing so, and thereby disrupt key helix-face protein–protein interactions necessary for DHFR activity.
A search of the available library of benzoylurea derivatives for an initial match yielded mimetics 1 and 2, that both fulfill the requirements of a hydrophobic face and the required phenylalanine substituent (Fig. 5A). Steady-state kinetic assays demonstrated that mimetic 1, which approximates an LxxFxxxF mimic, inhibited the enzyme with an IC50 of 800 µM for DHFR (Fig. 5B). Importantly, mimetic 2 (Fig. 5A), an FxxLxxxF mimic closer in sequence to the VxxLxxxF motif of the Beta Face residues, showed an increase in potency, with an IC50 of 230 µM (Fig. 5B, Table 2). The compound also inhibited TS, though to a lesser degree, as anticipated from the steady-state mutational data. This was not a non-specific effect, as other helix-mimetics with various side chains showed no inhibition of the enzyme at 1.5 mM. As expected from our mutational analysis, the mimetics, designed to maintain the interaction at the beta-sheet face, had no effect on the pre-steady-state rate of chemistry (data not shown).
Table 2.
IC50 values of mimetic 2 against enzyme targetsa
| Enzyme |
C. hominis DHFR |
C. hominis Beta Face DHFR |
Human DHFR |
C. hominis TS |
|---|---|---|---|---|
| IC50 (µM) | 230 ± 10 | 18 ± 1 | ≫1000 | 570 ± 60 |
All reactions were run with identical enzyme and ligand concentrations.
Values shown as mean ± standard deviation.
To delineate the mode of inhibition, we conducted a steady-state substrate profile for both DHFR substrates, H2folate and NADPH, in the presence and absence of mimetic 2. The data show that the α-helix mimetic acts as a noncompetitive (mixed) inhibitor for H2folate; Vmax decreased in the presence of inhibitor (1.46 ± 0.03 µmol min−1 in the absence of compound, and 0.48 ± 0.06 µmol min−1 in the presence of 250 µM compound) and the Km increased (0.8 ± 0.3 µM versus 13 ± 5 µM) (Fig. S4A†). The mimetic also acts as a noncompetitive inhibitor for NADPH; the Vmax decreased (1.33 ± 0.02 µmol min−1 in the absence of compound, and 0.41 ± 0.01 µmol min−1 in the presence of 250 µM compound), whereas the Km was not significantly altered (3.4 ± 0.4 µM versus 4.1 ± 1.1 µM) (Fig. S4A†). Lineweaver–Burk plots better demonstrate the noncompetitive character of the data (Fig. S4B†). As expected, the same noncompetitive mode of inhibition was obtained for mimetic 1 (data not shown).
To further confirm of the binding site of the α-helix mimetic, we ran steady-state inhibition assays against the Beta Face mutant enzyme, which no longer contains residues that would compete with the mimetic for the putative binding site. Mimetic 2 is at least 10-fold more potent against Beta Face than WT, with an IC50 of ~20 µM (Table 2). Additionally, the specificity of the lead mimetic was assessed by inhibition assays against the human enzyme. Importantly, there was no inhibition of human DHFR up to 1 mM mimetic 2, indicating species-specificity for the C. hominis enzyme (Table 2).
To examine if these inhibitors might exploit the defect in enzyme stability displayed by the crossover helix mutants and thus exhibit the dual effect of enzyme inhibition and destabilization, we wanted to examine the effect that the inhibitors might have on enzyme stability. While spectroscopic interference of the α-helix mimetics prevented the use of CD to monitor thermal stability, we were able to take advantage of our protease sensitivity assays as another readout of enzyme stability. As shown in Fig. 5C, mimetics 1 and 2 incubated at saturating concentrations with WT TS–DHFR, render the enzyme dramatically more sensitive to trypsin proteolysis. A side-by-side experiment with the known DHFR active site inhibitor methotrexate showed a resistance to proteolysis compared with the enzyme alone (Fig. 5C), confirming the general stabilizing effect of typical DHFR inhibitors.
Discussion
Insights into the role of the crossover helix for TS–DHFR function
Thymidylate synthase–dihydrofolate reductase (TS–DHFR) from C. hominis contains a unique DHFR domain in that the “crossover helix”, a swap domain between DHFR monomers, packs in a cleft at the backside of the DHFR active site. Here, we have used a kinetic and biophysical approach to demonstrate that the crossover helix mediates overall enzyme function. Disruption of crossover helix interactions with the DHFR domain severely impairs steady-state activity, pre-steady-state hydride transfer, and stability of the protein at physiological temperatures and in the presence of proteases. It is clear that the presence of an intact crossover helix is necessary for a fully active DHFR in this parasite.
The stability of the crossover helix mutant enzymes has interesting physiological implications for the role of the swap domain. It appears that all interactions of the crossover helix are necessary for stability, as even the F207A mutant enzyme showed a modest decrease in melting temperature. The important area of the melting curves to note is at physiological temperature, 37 °C. Whereas the WT enzyme is fully folded, all of the mutant enzymes are unstable. Moreover, all mutants except F207A are more sensitive to common proteases. These data all emphasize a role for the crossover helix in imparting stability to the enzyme under cellular conditions.
Our mutational analysis highlights the complexity with which DHFR catalysis is modulated by the crossover helix. While a complete crossover helix is necessary to impart enzyme stability, the various catalytic activities of the domains appear to be governed by separate faces of the helix. The enzymes in which the Helix Face of the crossover helix is disrupted (Ala Helix Face and All Alanine) show a roughly two-fold loss in DHFR activity. The enzymes in which the Beta Face of the crossover helix is disrupted (Ala Beta Face and All Alanine) show a two-fold loss in TS activity. Therefore, the crossover helix is able to modulate both enzymatic activities; interacting with Helix B to stabilize DHFR activity and with the DHFR beta-sheet to stabilize TS activity. Additionally, DHFR hydride transfer is unique from these effects as well, as disruption of any of the helix faces lead to a dramatic rate decrease, regardless of steady-state activity.
The most surprising mechanistic finding from this analysis is that this dramatic impairment in the rate of hydride transfer could be narrowed down to one key amino acid, Phe207. It is clear from the X-ray crystal structure of F207A that small but significant changes in the positioning of the crossover helix are the main effect of the mutant. Viewing the helix as a rigid body, Phe207 appears to work as an anchor that positions the helix in proper orientation for catalysis. Indeed, that mutation of the two interacting phenylalanines, which reside on the beta strand known to confer communication to the active site,20 show no decrease in activity supports the mechanism that Phe207 works not by modulating specific DHFR residues, but more globally by positioning the crossover helix in the correct orientation to the DHFR active site.
The fact that the mutant enzyme F207A affects only the pre-steady-state DHFR rate highlights the importance of a pre-steady-state or transient kinetic approach; simply focusing on the steady state rate would only have given information on the rate-limiting step of product release. The ability to monitor the rate of chemistry at the active site, specifically hydride transfer, has elucidated a more dramatic effect of mutating the crossover helix. Clearly the hydride transfer step is much more sensitive to changes in the position of the crossover helix than the steady-state rate. It is known that the M20 loop in E. coli DHFR fluctuates at rates comparable to steady-state turnover. It is also established that local fluctuations of the M20 loop in E. coli DHFR govern to some degree the transient catalytic rates along the DHFR reaction pathway.21 Our crystal structure suggests that proper positioning of the crossover helix maintains optimal interactions with Helix B, which is directly connected to the M20 loop. Together, a possible model of rate enhancement is that the crossover helix, when oriented properly, allows Helix B and the M20 loop to sample more productive conformations for the transient rates only, leading to an increased rate of hydride transfer.
This insight into the role of the crossover helix further differentiates C. hominis TS–DHFR from the other protozoan species. All protozoan parasitic bifunctional TS–DHFR enzymes (L. major, T. gondii, P. falciparum, and C. hominis) studied appear to maintain a DHFR kchem of ~150 s−1.4,22–24 It appears that regulation of catalytic function and domain interactions is highly species specific. While other species (L. major, P. falciparum) utilize interdomain ligand activation (TS-to-DHFR) and an N-terminal tail to enhance the DHFR catalytic rate, ChDHFR makes use of a helical swap domain to achieve the same effect.4,24 Additionally, while C. hominis TS–DHFR has not shown the particular type of TS-to-DHFR interdomain communication, the data presented here clearly indicate a role for the DHFR crossover helix in TS activity, the active site of which is 30 Å away from the swap domain. None of the amino acids of the crossover helix specifically contact the TS domain, however multiple amino acids of the returning linker, in sequence directly following the crossover helix, make specific contacts with non-active site regions of the TS domain. It is possible that the crossover helix modulates TS activity directly through the DHFR domain, or through disruption of the adjacent residues that contact the TS domain. Regardless of the physical mechanism, this implies that distal regions of the TS domain are important for ChTS catalysis. We have previously established that ChTS is unusually fast compared to other orthologs, one determinant of which was found to be unusual positioning of TS ligands.25,26 It will be interesting to determine if the crossover helix has a role in TS ligand orientation, or contributes to TS catalysis by another mechanism. Mutational and structural research is currently underway to pinpoint the effect of the crossover helix on the TS domain.
Novel inhibitor development
The use of α-helix mimetics to disrupt protein–protein interactions is an intriguing new chemical biology focus, combining the specificity of protein helical side chain projections and the stability and synthetic advantages of small molecules. We used an α-helix mimetic to serve the dual purpose of allowing us to further probe the mechanism of crossover helix enhancement, as well as to define a novel non-active site amenable to enzyme inhibition.
First, the kinetic pattern that we observed from incubation of the enzyme with the helix mimetic confirmed the results from our mutational analysis. Designed to disrupt only the Helix Face, we observed DHFR steady-state inhibition, with a lesser effect on TS, which is in line with the Ala Helix Face mutant enzyme data. Importantly, there was no decrease in the rate of hydride transfer. This helps to resolve a complex interpretation from the mutant analysis, and confirms that the Beta-Face, and most likely Phe207, are the crucial interactions mediating the transient rate of hydride transfer, and that the mechanism of rate enhancement is not directly through the interactions of the Helix Face.
The second notable result from this study was the ability of detailed biochemical data to guide the rational design of a helical mimetic inhibitor of an important protein–protein interaction and, as a result, enzyme activity. The activity data on the separate faces of the crossover helix confirmed the necessity to inhibit the helix–helix interaction, and the importance of Phe207 in docking the crossover helix led us towards a compound mimicking a phenylalanine at the third position. These insights allowed for the identification of a mid-micro-molar inhibitor of this novel cleft without the need for a high-throughput or virtual screening approach. Importantly, while peptidomimetics have proven useful for inhibition of enzymes such as proteases,27 and α-helix mimetics have shown recent promise for cancer drug targets via disruption of protein–protein interactions,28,29 to our knowledge this is the first report of an α-helix mimetic that combines both aspects; inhibiting the catalytic activity of an enzyme by disrupting a non-active site protein–protein interaction.
The fact that mimetic 2 shows selectivity for ChDHFR over the human enzyme validates this allosteric region as a potential site for novel therapeutics for the cryptosporidium parasite. While we view this mimetic as simply a proof-of-concept candidate for a lead compound, it seems likely that further synthetic design based on the structure of the ChDHFR helix cleft will yield considerably more potent inhibitors. The three amino acid stretch used here is not perfectly mimicked; the optimal VxxLxxxF mimic would likely yield greater potency, as well as substitute the smaller valine for the bulky valine, increasing ligand efficiency. Additionally, the binding cleft of the crossover helix has an overall negative electrostatic potential, making neutral or positively charged capping groups attractive for further inhibitor development (see Fig. S5 in ESI†). Again, this could be accomplished without an addition of molecular weight, and perhaps the ability to decrease size based on capping groups such as for instance an amide rather than carboxylate. This potential compound, with a matched side chain profile and electrostatic surface, is a target for a larger future effort to increase potency. An integrated approach of obtaining structural data of enzyme–inhibitor complexes, carrying out larger scale combinatorial synthesis for SAR, and utilizing computational modeling, all aimed at better elucidating the enzyme–inhibitor interactions crucial for optimal inhibitor activity, will be the most effective course to generate potent lead compounds for drug design efforts, and is currently underway.
The finding that we were able to destabilize the enzyme target is promising for further development of inhibitors for this region. Most small molecules that bind to enzymes lead to an increase in enzyme stability, by providing additional interactions and packing surfaces in otherwise flexible clefts.30 This has been specifically shown for both DHFR and TS.31,32 Indeed, methotrexate exerted a stabilization effect in our protease assays. However, the α-helix mimetics presented here clearly exert the opposite effect. The ability to simultaneously inhibit enzyme activity while rendering it more susceptible to proteolytic degradation represents a multifaceted approach to targeting ChTS–DHFR under cellular conditions. The few inhibitors that have disrupted enzyme stability, either directly or indirectly, have shown promise as lead compounds.33–35
Significance
The current study validates an allosteric region as crucial for overall function of ChTS–DHFR, an essential enzyme in the causative agent of cryptosporidiosis. Unique from its orthologs in humans and structurally similar protozoan parasites, precise packing of the crossover helix into a non-active site cleft of ChDHFR mediates both stability and multiple facets of catalytic activity in a complex manner than involves two different faces of the helix. Importantly, in using α-helical mimetics to probe the function of the crossover helix, we have identified a surface amenable to species-specific inhibitor design that at once allows for catalytic inhibition and enzyme destabilization. Given that there is no fully effective treatment for cryptosporidiosis, this species-unique region provides a starting point for the development of low-toxicity inhibitors to complement the available active site therapies.
More broadly, we have highlighted the importance of a detailed biochemical approach in dissecting a protein–protein interaction, and have demonstrated the complexity by which such an interaction can modulate enzyme activity. The fact that mutational data allowed for the rational prediction of activity disruption has clear implications for the promise of inhibitor development based on enzyme structure–function data when the benefit of a starting compound or scaffold is lacking.
Experimental procedures
Construction and expression of mutations ChTS–DHFR enzymes
Full length C. hominis TS–DHFR gene (derived from a human parasite clone), was encoded in the pTrc99A-rHCp, provided by Dr Richard G. Nelson and Dr Amy C. Anderson. Mutations to the crossover helix were made following the QuikChange mutagenesis kit protocol (Stratagene). Along with single-site and double mutants named by their amino acid mutations, the larger mutant enzymes constructed were: Ala Helix Face (K194A, D198A, L202A, D205A, I206A, and R210A); Ala Beta Face (S195A, I196A, T199A, V200A, L203A, F207A); and All Alanine (195–208 to alanines). Based on the secondary structure prediction software nnPredict, all of the mutants containing alanines were predicted to maintain helical character. WT and mutant enzymes were over-expressed in E. coli (BL21), grown at either 37 °C or 25 °C, and purified as previously described.
Circular dichroism spectroscopy
Enzymes at a final concentration of 10 µM were incubated in 1× reaction buffer (25 mM Tris pH 7.3, 25 mM MgCl2) were analyzed for CD absorption by an AVIV spectrophotometer Model 215 (AVIV Instruments, Inc.). Thermal denaturation was monitored by CD absorption at 222 nm. The sample was cooled to 4 °C and a thermal scan was initiated, measuring absorption at 1 °C intervals to 90 °C, with an equilibration time of 60 seconds per step. The baseline for folded (low temperature) and unfolded (high temperature) protein was determined by averaging the values from 4–15 °C and 79–90 °C, respectively, and the fraction folded was calculated. The melting temperature was determined by fitting the data to a single-step transition.
Protease sensitivity
Enzymes at a final concentration of 10 µM were incubated in protease buffer (50 mM Tris, pH 8.0). Either trypsin or Glu-C was added to the reaction at a final w/w protease/TS–DHFR ratio of 1/500 and 1/250, respectively. Aliquots of 20 µL of the protease reactions were quenched with SDS loading buffer (50 mM Tris pH 8.0, 5 mM EDTA, 10% glycerol, 5% β-mercaptoethanol, 2% SDS, 0.02% bromophenol blue) at either 1, 15, or 60 minutes, and boiled for 5 minutes. The zero time-point was made by adding reaction mix without protease to SDS loading buffer, and then adding the proper concentration of protease before boiling. Samples were run on a 15% SDS polyacrylamide gel, and stained with SimplyBlue SafeStain (Invitrogen). Proteolysis was quantitated using a Molecular Imager FX (BioRad), calculating the density of the band corresponding to the full-length enzyme, and normalizing to the zero time-point.
Kinetic analysis of mutant enzymes
Steady-state kinetic analysis was conducted for each individual reaction (DHFR and TS) by spectroscopy at 340 nm, as previously described.22 The steady-state rate was determined by converting specific activity (µmol min−1 mg−1 protein) to a turnover number (kcat) using the active enzyme concentration in the reaction. Pre-steady-state kinetics were conducted using a stopped-flow fluorescence assay on a Kintek SF-2001 apparatus (Kintek Instruments, Austin TX). The assay utilizes the coenzyme fluorescence resonance energy transfer to NADPH to monitor the rate of DHFR chemistry. Excitation at 287 nm is absorbed by the enzyme, the emission at 340 nm is absorbed by NADPH, and the resulting emission is measured as fluorescence using an output filter of 450 nm. As NADPH is consumed, the emission signal will decrease as a function of the rate of the reaction. Single-turnover experiments to measure the rate of chemistry at the active site were conducted, with enzyme (50 µM) incubated with NADPH (500 µM), and then rapidly mixed with 10 µM H2folate. Data were collected as 1000 observation points over 50 ms (fast enzymes) or 300 ms (slow enzymes). The data were fit to a single exponential equation to obtain the DHFR rate constant.
X-ray crystal structure of F207A mutant enzyme
Pure ChTS–DHFR F207A mutant protein was incubated at a final concentration of 7 mg mL−1 with 1 mM ligands (dUMP, CB3717, NADPH, and methotrexate) for 45 min on ice and crystallized by hanging drop vapor diffusion. The successful well solution consisted of 0.04 M ammonium sulfate, 0.12 M lithium sulfate, 0.1 M Tris, and 16% polyethylene glycol 6000. Crystals with approximate dimensions of 0.05 mm × 0.05 mm × 0.1 mm grew in 1 week at 18 °C. Crystals were soaked in successive cryoprotectants of mother liquor with 10, 15 and 20% ethylene glycol for 1 min each and were flash-frozen in liquid nitrogen. Diffraction data were collected at the Brookhaven National Laboratory on beamline X29. Our best crystal diffracted to 2.6 Å. Data were indexed, integrated, and scaled to 2.7 Å using HKL2000 in a trigonal space group, and were converted to structure factors with truncate, with five percent of the reflections marked for cross-validation analysis to serve as Rfree. Phasing was solved by molecular replacement using Amore, with the coordinates of wild-type ChTS–DHFR (Protein Data Bank entry 1QZF), with waters and ligands removed, as the search model. The molecular replacement search confirmed a space group of P3221, and indicated four monomers (two dimers) in the asymmetric unit. After a rigid-body fit using Refmac5 from the CCP4 suite, the initial R-factor was 38.8%. The negative difference density for the absent Phe207 side chain was clear at 3s in every monomer in the initial difference density map. Additionally, DHFR ligands were visible in the initial Fo − Fc difference maps. The structure was refined using Refmac5;36 density modification was conducted using Solomon,37 and refinement and manual residue and ligand positioning were carried out in the visualization program COOT.38 After addition of waters in Refmac5, group B-factor refinement in CNS,39 and geometry optimization, the final Rfac was 22.2 and Rfree was 27.3. All refinement statistics are reported in Table 1. The structure is deposited in the Protein Data Bank as entry PDB 3HJ3.
Analysis of α-helix mimetics
Benzoylurea derivatives that mimicked crossover helix side chain projections were synthesized as described previously.19 Steady-state inhibition assays were conducted as above, with the addition that the helix mimetic was incubated with enzyme and substrates for 30 minutes prior to initiation of the reaction. As the compounds were brought up to stock concentrations in DMSO, the same volume of DMSO was added as a control for the reaction without inhibitor, and all rates were normalized to this “DMSO blank”. For determination of mode of inhibition, assays were conducted with varying substrate concentration, and the rate profile and Lineweaver–Burk plot were constructed and analyzed using standard Michaelis–Menten treatment. Protease sensitivity experiments were conducted as above, with a near-saturating concentration of helix-mimetic or methotrexate, incubated for 15 minutes prior to initiation of the reaction.
Supplementary Material
Acknowledgements
We would like to thank the entire staff at Beamline X29 at Brookhaven National Labs for their gracious technical support, and Dr Gregg Crichlow for crystallographic discussions. This work was supported in part by NIH Grant 5T32-AI 07404 (to W. E. M.), NIH Grant GM 69850 (to A. D. H.), and NIH Grants AI 44630 and AI 083146 (to K. S. A).
Footnotes
TS–DHFR, thymidylate synthase–dihydrofolate reductase is a functional designation as catalysis at TS precedes catalysis at DHFR; elsewhere the bifunctional enzyme is called DHFR-TS based on DHFR being N-terminal to TS.
References
- 1.Ivanetich KM, Santi DV. Bifunctional thymidylate synthase–dihydrofolate reductase in protozoa. FASEB J. 1990;4(6):1591–1597. doi: 10.1096/fasebj.4.6.2180768. [DOI] [PubMed] [Google Scholar]
- 2.O'Neil RH, et al. Phylogenic classification of protozoa based on the structure of the linker domain in the bifunctional enzyme, dihydrofolate reductase–thymidylate synthase. J. Biol. Chem. 2003;278:52980–52987. doi: 10.1074/jbc.M310328200. [DOI] [PubMed] [Google Scholar]
- 3.Yuvaniyama J, et al. Insights into antifolate resistance from malarial DHFR–TS structures. Nat. Struct. Biol. 2003;10(5):357–365. doi: 10.1038/nsb921. [DOI] [PubMed] [Google Scholar]
- 4.Dasgupta T, Anderson KS. Probing the role of parasite-specific, distant structural regions on communication and catalysis in the bifunctional thymidylate synthase–dihydrofolate reductase from Plasmodium falciparum. Biochemistry. 2008;47(5):1336–1345. doi: 10.1021/bi701624u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vargo MA, Martucci WE, Anderson KS. Disruption of the crossover helix impairs dihydrofolate reductase activity in the bifunctional enzyme TS–DHFR from Cryptosporidium hominis. Biochem. J. 2009;417(3):757–764. doi: 10.1042/BJ20081247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mead JR. Cryptosporidiosis and the challenges of chemotherapy. Drug Resist. Updates. 2002;5:47–57. doi: 10.1016/s1368-7646(02)00011-0. [DOI] [PubMed] [Google Scholar]
- 7.Tzipori S, Widmer G. A hundred-year retrospective on cryptosporidiosis. Trends Parasitol. 2008;24(4):184–189. doi: 10.1016/j.pt.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dillingham R, Lima A, Guerrant R. Cryptosporidiosis: epidemiology and impact. Microbes Infect. 2002;4(10):1059–1066. doi: 10.1016/s1286-4579(02)01630-1. [DOI] [PubMed] [Google Scholar]
- 9.Gilles HM, Hoffman PS. Treatment of intestinal parasitic infections: a review of nitazoxanide. Trends Parasitol. 2002;18(3):95–97. doi: 10.1016/s1471-4922(01)02205-x. [DOI] [PubMed] [Google Scholar]
- 10.Bolstad DB, et al. Structure-based approach to the development of potent and selective inhibitors of dihydrofolate reductase from cryptosporidium. J. Med. Chem. 2008;51(21):6839–6852. doi: 10.1021/jm8009124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brophy VH, et al. Identification of Cryptosporidium parvum dihydrofolate reductase inhibitors by complementation in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 2000;44(4):1019–1028. doi: 10.1128/aac.44.4.1019-1028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson RG, Rosowsky A. Dicyclic and tricyclic diaminopyrimidine derivatives as potent inhibitors of Cryptosporidium parvum dihydrofolate reductase: structure–activity and structure–selectivity correlations. Antimicrob. Agents Chemother. 2001;45(12):3293–3303. doi: 10.1128/AAC.45.12.3293-3303.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martucci WE, et al. Novel non-active site inhibitor of Cryptosporidium hominis TS–DHFR identified by a virtual screen. Bioorg. Med. Chem. Lett. 2009;19(2):418–423. doi: 10.1016/j.bmcl.2008.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dwyer DS. Electronic properties of the amino acid side chains contribute to the structural preferences in protein folding. J. Biomol. Struct. Dyn. 2001;18(6):881–892. doi: 10.1080/07391102.2001.10506715. [DOI] [PubMed] [Google Scholar]
- 15.Kneller DG, Cohen FE, Langridge R. Improvements in protein secondary structure prediction by an enhanced neural network. J. Mol. Biol. 1990;214(1):171–182. doi: 10.1016/0022-2836(90)90154-E. [DOI] [PubMed] [Google Scholar]
- 16.Kosinski MJ, Bailey JE. Structural characteristics of an abnormal protein influencing its proteolytic susceptibility. J. Biotechnol. 1992;23(2):211–223. doi: 10.1016/0168-1656(92)90093-o. [DOI] [PubMed] [Google Scholar]
- 17.Magagnoli C, et al. Mutations in the A subunit affect yield, stability, and protease sensitivity of nontoxic derivatives of heat-labile enterotoxin. Infect. Immun. 1996;64(12):5434– 5438. doi: 10.1128/iai.64.12.5434-5438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saxl RL, et al. Significance of mutations on the structural perturbation of thymidylate synthase: implications for their involvement in subunit exchange. Protein Sci. 2007;16(7):1439–1448. doi: 10.1110/ps.062509807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez JM, Hamilton AD. Benzoylurea oligomers: synthetic foldamers that mimic extended alpha helices. Angew. Chem., Int. Ed. Engl. 2007;46(45):8614–8617. doi: 10.1002/anie.200701869. [DOI] [PubMed] [Google Scholar]
- 20.Dion A, et al. How do mutations at phenylalanine-153 and isoleucine-155 partially suppress the effects of the aspartate-27 → serine mutation in Escherichia coli dihydrofolate reductase? Biochemistry. 1993;32(13):3479–3487. doi: 10.1021/bi00064a036. [DOI] [PubMed] [Google Scholar]
- 21.McElheny D, et al. Defining the role of active-site loop fluctuations in dihydrofolate reductase catalysis. Proc. Natl. Acad. Sci. U. S. A. 2005;102(14):5032–5037. doi: 10.1073/pnas.0500699102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atreya CE, Anderson KS. Kinetic Characterization of Bifunctional Thymidylate Synthase–Dihydrofolate Reductase (TS–DHFR) from Cryptosporidium hominis: A Paradigm Shift for TS Activity and Channeling Behavior. J. Biol. Chem. 2004;279:18314–18322. doi: 10.1074/jbc.M400009200. [DOI] [PubMed] [Google Scholar]
- 23.Johnson EF, et al. Mechanistic characterization of Toxoplasma gondii thymidylate synthase (TS–DHFR)–dihydrofolate reductase. Evidence for a TS intermediate and TS half-sites reactivity. J. Biol. Chem. 2002;277(45):43126–43136. doi: 10.1074/jbc.M206523200. [DOI] [PubMed] [Google Scholar]
- 24.Liang P-H, Anderson KS. Kinetic reaction scheme for the dihydrofolate reductase domain of the bifunctional thymidylate synthase–dihydrofolate reductase from Leishmania major. Biochemistry. 1998;37:12206–12212. doi: 10.1021/bi9803170. [DOI] [PubMed] [Google Scholar]
- 25.Doan LT, et al. Nonconserved residues Ala287 and Ser290 of the Cryptosporidium hominis thymidylate synthase domain facilitate its rapid rate of catalysis. Biochemistry. 2007;46(28):8379–8391. doi: 10.1021/bi700531r. [DOI] [PubMed] [Google Scholar]
- 26.Martucci WE, Vargo MA, Anderson KS. Explaining an unusually fast parasitic enzyme: folate tail-binding residues dictate substrate positioning and catalysis in Cryptosporidium hominis thymidylate synthase. Biochemistry. 2008;47(34):8902–8911. doi: 10.1021/bi800466z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen JT, et al. Design of potent aspartic protease inhibitors to treat various diseases. Arch. Pharm. 2008;341(9):523–535. doi: 10.1002/ardp.200700267. [DOI] [PubMed] [Google Scholar]
- 28.Avendano C, Menendez JC. Peptidomimetics in cancer chemotherapy. Clin. Transl. Oncol. 2007;9(9):563–570. doi: 10.1007/s12094-007-0104-6. [DOI] [PubMed] [Google Scholar]
- 29.Davis JM, Tsou LK, Hamilton AD. Synthetic non-peptide mimetics of alpha-helices. Chem. Soc. Rev. 2007;36(2):326–334. doi: 10.1039/b608043j. [DOI] [PubMed] [Google Scholar]
- 30.Cimmperman P, et al. A quantitative model of thermal stabilization and destabilization of proteins by ligands. Biophys. J. 2008;95(7):3222–3231. doi: 10.1529/biophysj.108.134973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ainavarapu SR, et al. Ligand binding modulates the mechanical stability of dihydrofolate reductase. Biophys. J. 2005;89(5):3337–3344. doi: 10.1529/biophysj.105.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berger SH, Berger FG, Lebioda L. Effects of ligand binding and conformational switching on intracellular stability of human thymidylate synthase. Biochim. Biophys. Acta. 2004;1696(1):15–22. doi: 10.1016/j.bbapap.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Sekine K, et al. Small molecules destabilize cIAP1 by activating auto-ubiquitylation. J. Biol. Chem. 2008;283(14):8961–8968. doi: 10.1074/jbc.M709525200. [DOI] [PubMed] [Google Scholar]
- 34.Todd MJ, Freire E. The effect of inhibitor binding on the structural stability and cooperativity of the HIV-1 protease. Proteins: Struct., Funct., Genet. 1999;36(2):147–156. doi: 10.1002/(sici)1097-0134(19990801)36:2<147::aid-prot2>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Chen S. Aromatase destabilizer: novel action of exemestane, a food and drug administration-approved aromatase inhibitor. Cancer Res. 2006;66(21):10281–10286. doi: 10.1158/0008-5472.CAN-06-2134. [DOI] [PubMed] [Google Scholar]
- 36.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 37.Collaborative Computational Project. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. No. 4. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Brunger AT, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.