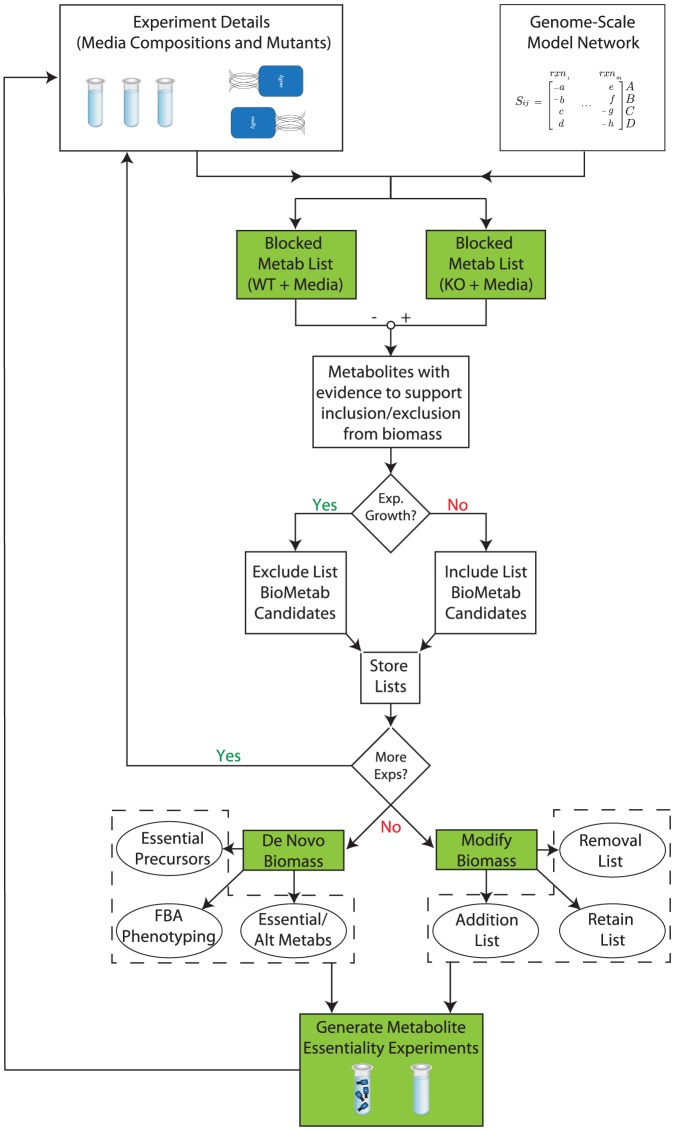
Figure 1. A flow diagram summarizing the BioMog framework.
Initially, mutants are evaluated computationally to determine metabolites that are blocked in mutant strains but not in the wild type strain. While not strictly necessary, mutants that have substantially deleterious impacts on the network (>100 blocked metabolites) are considered uninformative and filtered out to improve the quality of proposed biomass components. Blocked metabolites are then assigned to an include/exclude metabolite list based on the experimentally observed growth phenotype. Once these lists have been created, BioMog can use this information to propose de novo biomass components or to modify the existing biomass equation. Additional experiments can be designed and run by BioMog to fill in any informational gaps that may exist in the current dataset after which the cycle can be repeated.