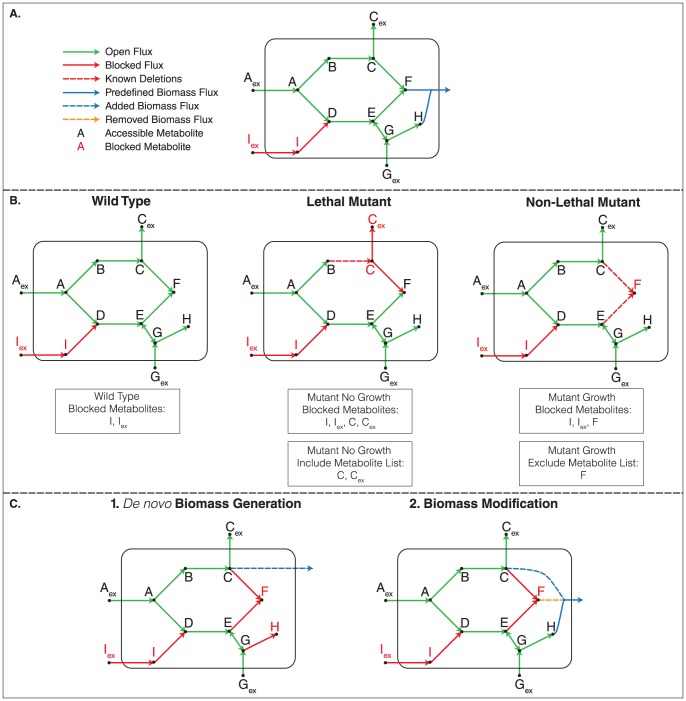
Figure 2. Application of BioMog to an illustrative example.
(A) For an existing model and set of biomass requirements (metabolites F and H), BioMog is capable (depending on the quality and quantity of data) of generating, de novo, an organism's biomass requirements or of modifying a predefined biomass equation. This is accomplished by removing the initial biomass equation from the network and adding sinks for every metabolite (not shown). Blocked metabolites are determined for the wild type and mutant strains under a particular experimental condition (B). This process is repeated for all growth phenotype experiments for which data exist. The set of blocked metabolites can then be used to propose a new biomass equation or modify an existing one (C). Based on the include/exclude metabolite lists generated in this example, the original biomass equation composed of substrates F and H is modified by adding metabolite C while removing F. Since the de novo biomass relies solely on experimental evidence, it is important that enough data exist that test the essentiality of different metabolites if one desires an accurate and complete understanding of the biomass requirements. Here, metabolite H was absent from the proposed de novo biomass because no supporting or refuting evidence existed in the experimental dataset to justify its inclusion.