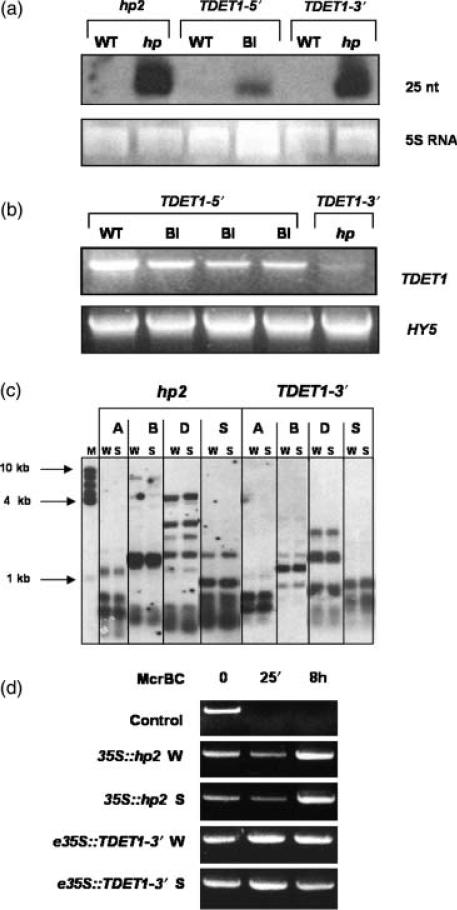
Figure 6.
Molecular analysis of TDET1 gene silencing.
(a) siRNA analysis of transgenic lines overexpressing different TDET1 constructs. In vitro-synthesized TDET1 was used as probe. 5S rRNA is shown as loading control.
(b) RT-PCR analysis of transgenic lines containing e35S::TDET1 5′ and e35S:: TDET1 3′ constructs. Full-length TDET1 was synthesized using specific TDET1 primers and HY5 primers were used as control for cDNA synthesis and loading.
(c) Methylation analysis of genomic DNA from transgenic plants containing 35S::hp2 and e35S::TDET1 3′ constructs. Genomic DNA from wild type (W) and fully silenced (S) plants was digested with Alu1 (A), BglII/EcoRI (B), Dde1 (D) and Sau3A1 (S). Full-length TDET1 was used as probe. 1 kb size markers are indicated.
(d) McrPCR analysis of genomic DNA from transgenic plants containing 35S::hp2 and e35S::TDET1 3′ constructs. Genomic DNA from wild type (W) and fully silenced (S) plants was digested with McrBC at the indicated times and the TDET1 transgene subsequently amplified by PCR. A methylated control DNA is also shown.
35S::hp2 plants were MM genotype, whereas the e35S::TDET1-3′ plants were T56 genotype. The DNA used for methylation analysis was derived from plants from T4 generation, and results obtained were identical to those found in the T3 generation (data not shown).