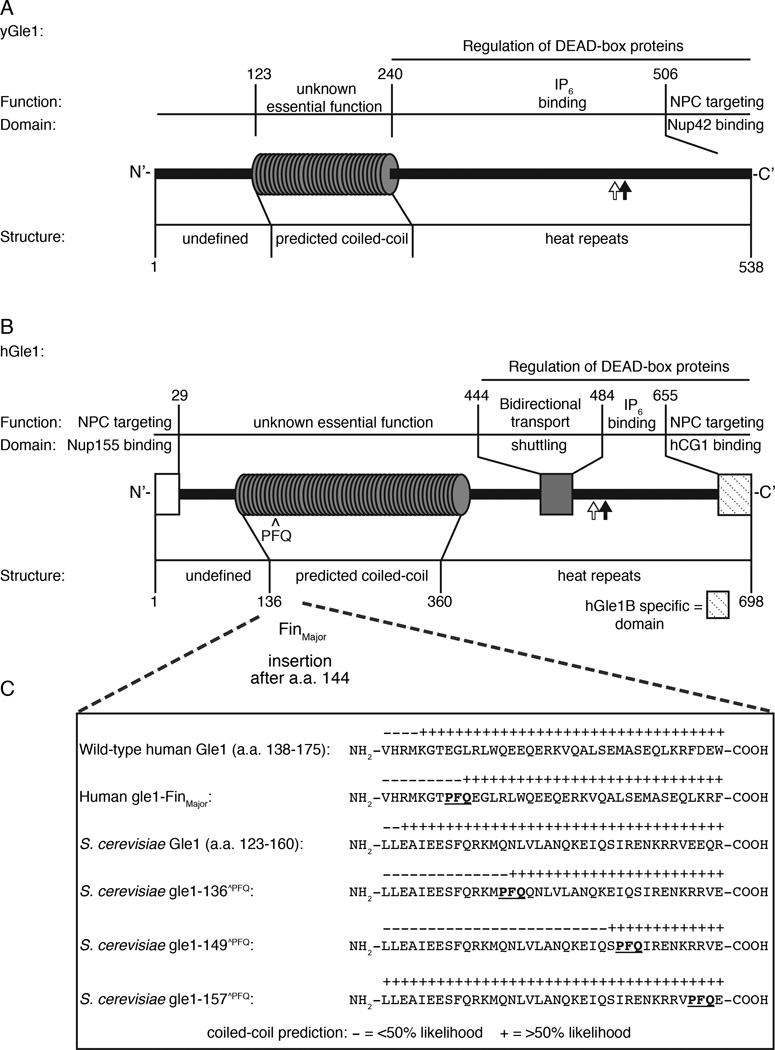
Figure 1. Conserved structural and functional elements of Gle1 from S. cerevisiae and humans.
(A) Diagram depicting functional and structural domains in S. cerevisiae (y)Gle1. Black arrow represents the position of the y-gle1-4 mutation (G382R) (Watkins et al., 1998). White arrow marks the location of conserved IP6-coordinating residues K377 and K378 (Alcazar-Roman et al., 2010). (B) Diagram depicting functional and structural domains in human (h)Gle1B (adapted from Kendirgi et al., 2005). “PFQ” denotes location of the FinMajor insertion after amino acid 144 in the predicted coiled-coil domain (Nousiainen et al., 2008). Black arrow represents the homologous position (Q548) of the residue mutated in y-gle1-4 (Watkins et al., 1998). White arrow marks the location of conserved IP6-coordinating residues K526 and K527 (Alcazar-Roman et al., 2010). (C) Paircoil2-generated structure predictions of Gle1 polypeptide sequences, showing the structural effect of h-gle1-FinMajor and y-gle1 engineered PFQ insertions. (+) indicates >50% probability of coiled-coil structure. PFQ insertions locations are designated by bold underlined typeface. See Table S1 and S2 for Paircoil2 scores and p-values.