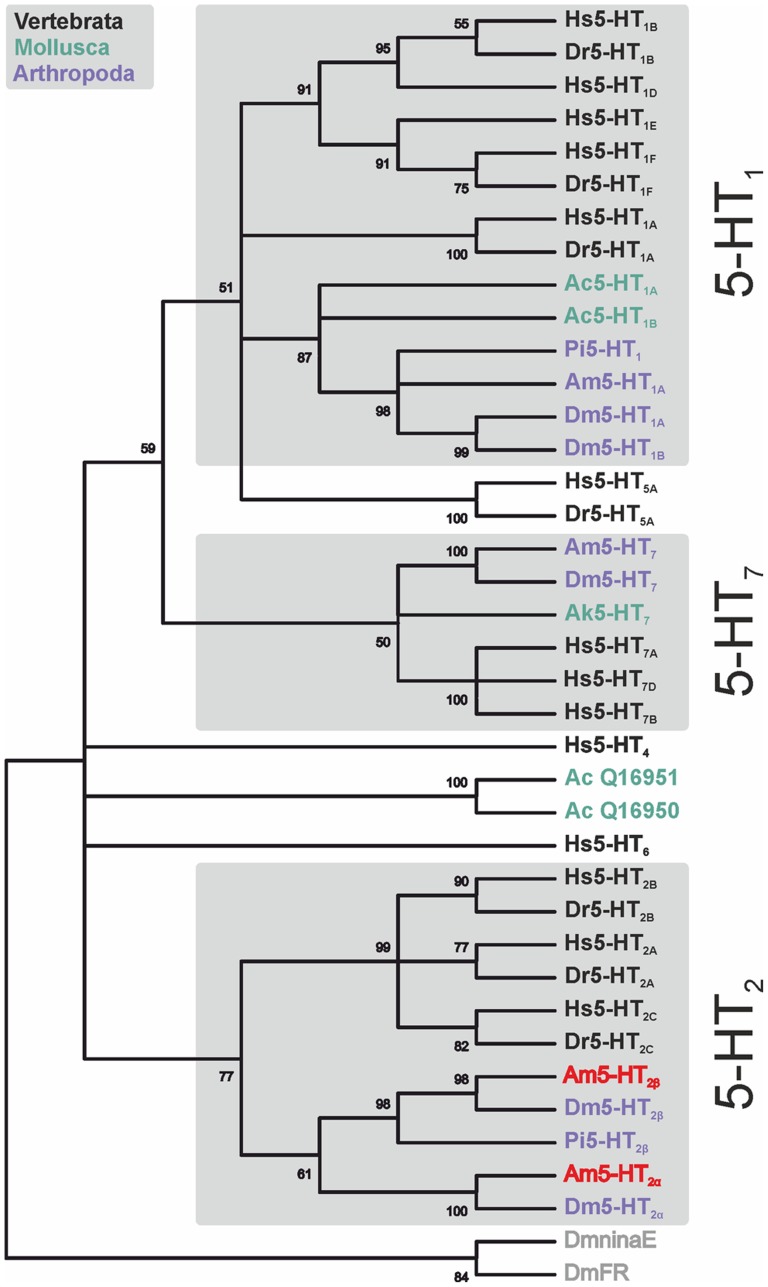
Figure 3. Phylogenetic relationship between various 5-HT receptors from vertebrates, mollusks, and arthropods.
Alignments were performed with BioEdit (version 7.0.5) by using the core amino-acid sequences lacking the variable regions of the N- and C-terminus and the third cytoplasmic loop. Maximum parsimony analysis was calculated with MEGA4. The receptor sequences followed by their accession numbers are listed in the order illustrated: Homo sapiens (Hs5-HT1B, acc. no. NP_000854), Danio rerio (Dr5-HT1B, AAI63698), Hs5-HT1D (NP_000855), Hs5-HT1E (NP_000856), Hs5-HT1F (NP_000857), Dr5-HT1F (NP_001076425), Hs5-HT1A (NP_000515), Dr5-HT1A (NP_001139238), Aplysia californica (Ac5-HT1, AAM46088, AAC28786), Panulirus interruptus (Pi5-HT1, AY528822), Apis mellifera (Am5-HT1, CBI75449), Drosophila melanogaster (Dm5-HT1A, CAA77570), Dm5-HT1B (CAA77571), Hs5-HT5A (NP_076917), Dr5-HT5A, (NP_001119882), Am5-HT7 (AM076717), Dm5-HT7 (NP_524599), Aplysia kurodai (Ak5-HT7, ACQ90247), Hs5-HT7A (NP_000863), Hs5-HT7D (NP_062873), Hs5-HT7B (NP_062874), Hs5-HT4 (NP_001035259), Ac5-HT2 (Q16951), Ac5-HT1 (Q16950), Hs5-HT6 (NP_000862), Hs5-HT2B (NP_000858), Dr5-HT2B (ABI18978), Hs5-HT2A (NP_000612), Dr5-HT2A (CAQ15355), Hs5-HT2C (NP_000859), Dr5-HT2C (CAX14715), Am5-HT2β (FR727108), Dm5-HT2β (NM_141548/NM_169229), Pi5-HT2β (AY550910), Am5-HT2α (FR727107), and Dm5-HT2α (CAA57429). The divergent D. melanogaster ninaE-encoded rhodopsin 1 (DmninaE, NM_079683) and D. melanogaster FMRFamide receptor (DmFR, AAF47700) were used as out-groups. The numbers at the nodes of the branches represent the percentage bootstrap support (2,000 replications) for each branch.