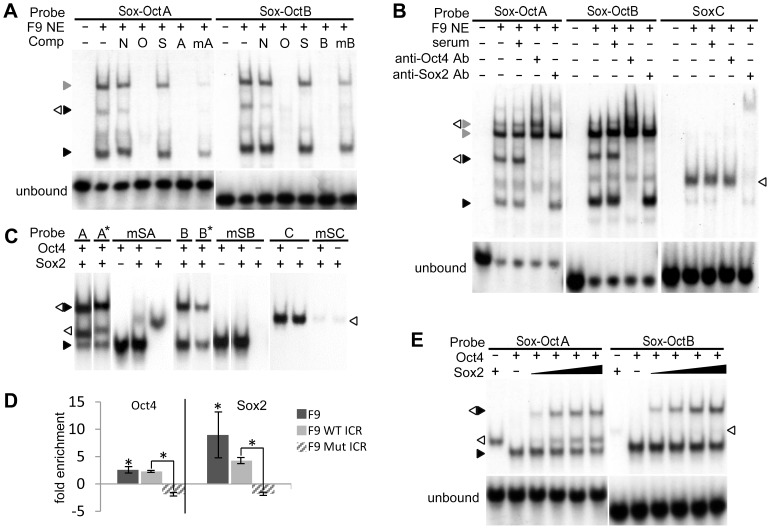
Figure 2. Sox2 and Oct4 binding to the ICR.
EMSAs were performed using WT or mutant ICR probes as indicated with either F9 NE (A, B) or recombinant Oct4 and Sox2 (C, D, F). (A) The effect of octamer specific point mutations on Oct4 and Sox2 binding was assessed by addition of nonspecific (N) and specific unlabeled DNA competitors - oct (O), sox (S), ICR Sox-OctA and B (A and B), or mutant ICR Sox-OctA and B (mA and mB). (B) Oct4 and Sox2 supershift assays. Sox2 and Oct4 protein-DNA complexes (white and black triangles respectively) were identified by their respective antibodies (Ab). Presumptive Oct1 complexes are indicated by a grey triangle. (C) Recombinant Sox2 binding to Sox-OctA, B and SoxC. ICR probes with mutations to the Sox elements (mSA, mSB, and mSC) were used to confirm Sox2 binding sites. (D) Methylation-independent binding of Oct4 and Sox2 to the ICR Sox-Oct motifs Wild type ICR Sox-OctA and B probes (A* and B*) were methylated at all CpGs. (E) Oct4 and Sox2 ChIP assays. Enrichment of endogenous F9 or WT and mutant transgenic ICR octamers by Oct4 and Sox2 ChIP was determined by qRT-PCR and normalized to serum controls. Error bars represent standard deviation of the mean (n = 3). Statistical significance of Oct4 and Sox2 enrichments was shown using Student's t-test (*p<0.05). (F) Cooperative binding of Sox2 to Oct4 complexes. Cooperative binding characteristics of Sox2 to the ICR were assessed by measuring the amount of binary and ternary protein-DNA complexes formed using a fixed amount of Oct4 and increasing amounts of Sox2. See Table S1 for a complete list of probe and competitor sequences.