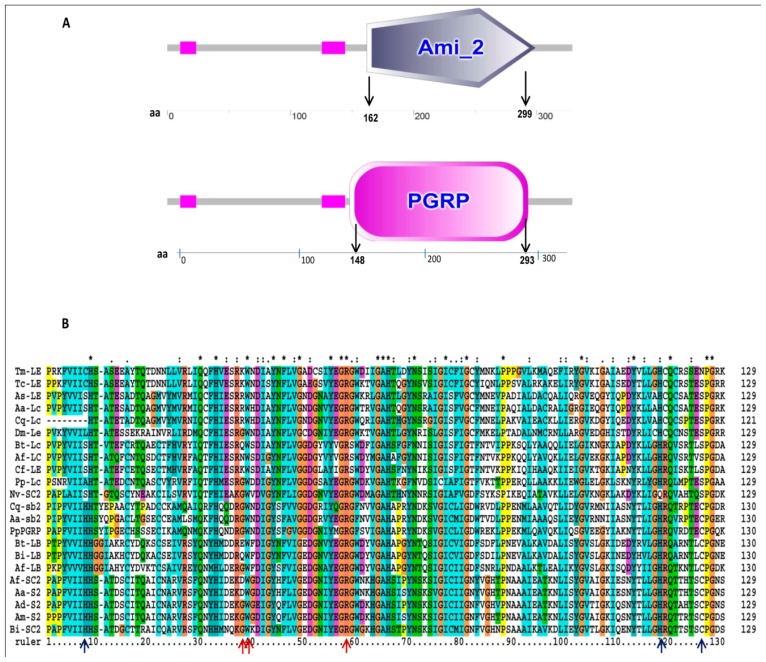
Figure 2.
Characterization and alignment of amino acid sequences of TmPGRP-LE homologues. (A) Domain architecture predicted using SMART analysis tool. Ami-2 and PGRP stand for predicted amidase-2 and PGRP domains, respectively; and (B) Multiple sequence alignment of amidase/PGRP domains of PGRP family members in representative insects. The sequences have been compiled from NCBI database and have been represented by accession numbers (Figure 3 for details). Catalytic residues for amidase activity and specific PGN recognition residues are indicated by blue and red arrows, respectively. Residues shared are indicated with the following symbols: “*” (denotes identical residues in all sequences); “:” (conserved substitutions according to similar properties of amino acids); “.” (indicates semi-conserved amino acid substitutions). Dashes indicate gaps used to maximize the alignment.