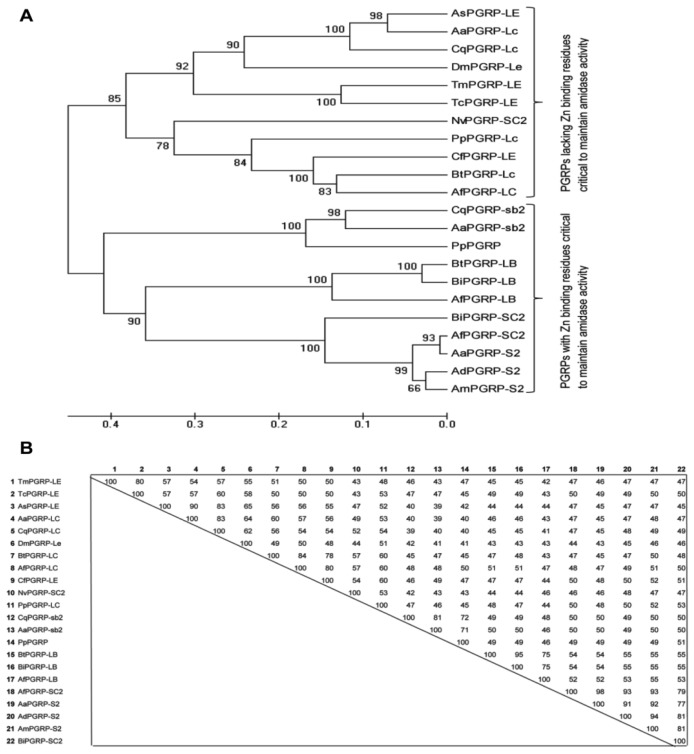
Figure 3.
Molecular phylogenetic and percentage identity analysis of TmPGRP-LE (A) Phylogenetic analysis as performed by MEGA 5.0 software with representative PGRP gene family members. The percentage of replicate trees in which the associated taxa clustered together in bootstrap test (5000 replicates) is shown on interior branches. The abbreviations and accession numbers of the sequences were as follows: PGRP-LE: Tribolium castaneum (TcPGRP-LE; XP_968926.1), Armigeres subalbatus (AsPGRP-LE; AEX31482.1), Drosophila melanogaster (DmPGRP-LEa; NP_573078.1), Camponotus floridanus (CfPGRP-LE; EFN63542.1); PGRP-LC: Aedes aegypti (AaPGRP-LC; XP_001656352.1), Culex quinquefasciatus (CqPGRP-LC; XP_001842237.1), Bombus terrestris (BtPGRP-LC; XP_003396511.1), Apis florea (AfPGRP-LC; XP_003693124.1); PGRP-SC2: Bombus impatiens (XP_003396511.1), Nasonia vitripennis (NvPGRP-SC2; XP_001603488.1), Apis florea (AfPGRP-SC2; XP_003694493.1); PGRP-SB2: Culex quinquefasciatus (CqPGRP-SB2; XP_001849091.1), Aedes aegypti (AaPGRP-SB2; XP_001654275.1); PGRP-LB: Bombus terrestris (BtPGRP-LB; XP_003400160.1), Bombus impatiens (BiPGRP-LB; XP_003493260.1), Apis florea (AfPGRP-LB; XP_003694545.1); PGRP-S2: Apis mellifera (AmPGRP-S2; NP_001157188.1), Apis dorsata (AdPGRP-S2; ACT66892.1); Phlebotomus papatasi (PpPGRP; ABV60369.1); and (B) Percentage identity matrix of the representative species as inferred using ClustalX 1.83.