Figure 5.
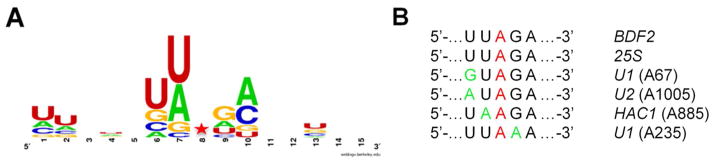
Analysis of yeast substrates reveals 3′ next nearest neighbor preference for adenosine. A WebLogo analysis of the hADAR2 edited transcripts in INVSc1 identified in the RNA-Seq. screen.(32) The sequence logo incorporates seven nucleotides 5′ and 3′ to the 19 edited sites from Table 1. B Alignment of six hADAR2 substrates found in S. cerevisiae closely related by sequence flanking the edited A. Red indicates editing site. Green indicates deviation from the sequence 5′-UUAGA-3′.
