Figure 2. Identification of PARN, poly(A)-specific ribonuclease, as the pre-miR-451 trimming enzyme.
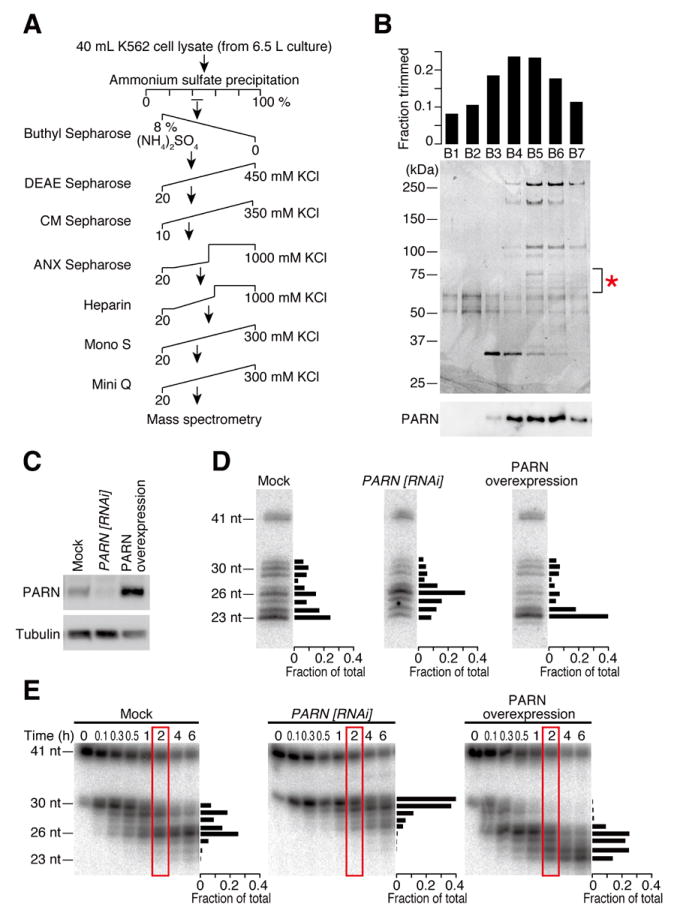
(A) Scheme of the chromatographic steps for purification of PARN.
(B) The profile of the final Mini Q purification. Each fraction was assayed for the trimming activity of ac-pre-miR-451 in vitro (top) and the proteins were separated by SDS-PAGE (middle). Protein bands that showed close correlation with the trimming activity were analyzed by mass spectrometry (indicated by *), among which PARN was found. Bottom, anti-PARN Western blotting.
(C) Anti-PARN Western blotting analysis of PARN knockdown or overexpression.
(D) Effect of PARN on pre-miR-451 trimming in HeLa S3 cells. Upon depletion or overexpression of PARN, a plasmid that expresses a miR-144/451 primary transcript was transfected and miR-451 was detected by Northern blotting. The signal intensities of the bands at 2 h are quantified and plotted on the right of each gel. PARN depletion impaired pre-miR-451 trimming while PARN overexpression promoted it. See also Figure S2 for miR-144 and endogenous let-7 controls.
(E) Effect of PARN on pre-miR-451 trimming in vitro. Lysates from PARN-depleted or PARN-overexpressing cells were tested for trimming. The signal intensities of the bands at 2 h (in red rectangles) are quantified and plotted on the right of each gel.
