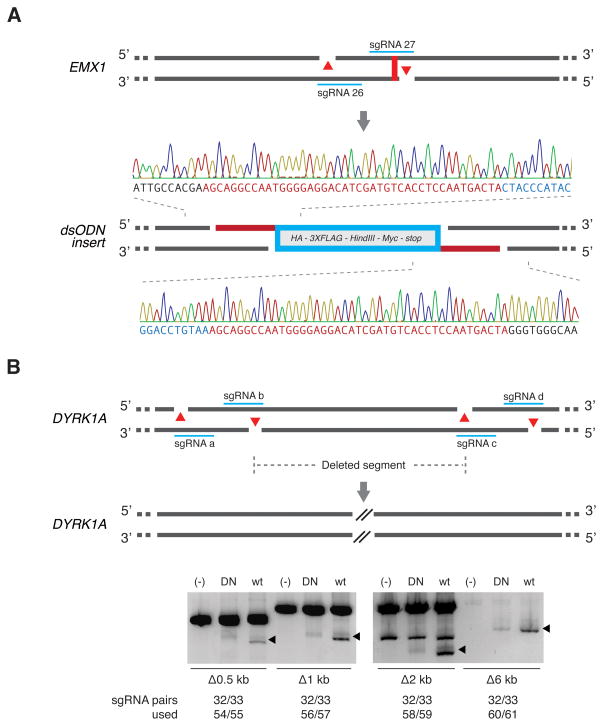
Figure 5. Multiplexed nicking facilitates non-HR mediated gene integration and genomic deletions.
(A) Schematic showing insertion of a double-stranded oligodeoxynucleotide (dsODN) donor fragment bearing overhangs complementary to 5′ overhangs created by Cas9 double nicking. The dsODN was designed to remove the native EMX1 stop codon and contains a HA tag, 3X FLAG tag, HindIII restriction site, Myc epitope tag, and a stop codon in frame, totaling 148 bp. Successful insertion was verified by Sanger sequencing as shown (1/37 clones screened). (B) Co-delivery of four sgRNAs with Cas9n generate long-range genomic deletions in the DYRK1A locus (from 0.5 kb up to 6 kb).