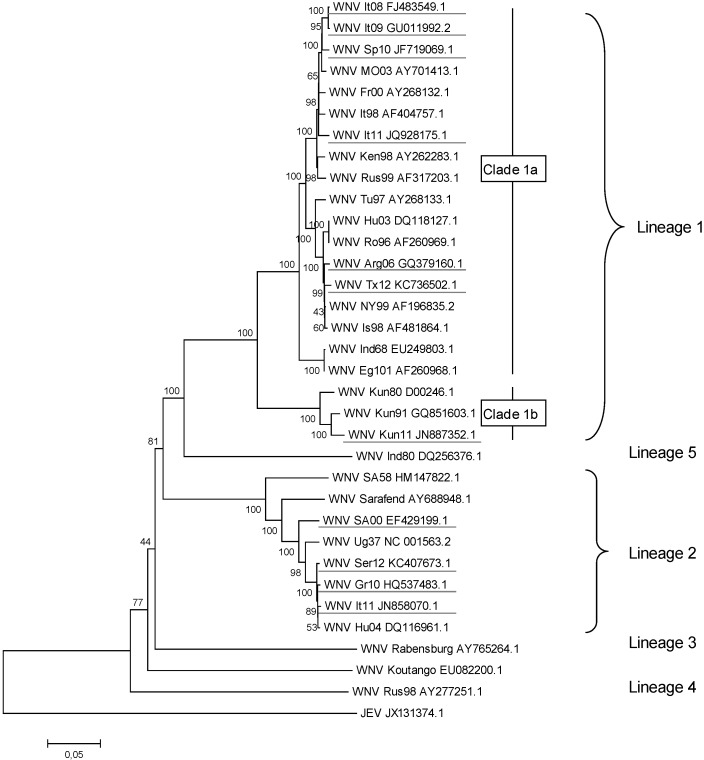
Figure 1.
West Nile Virus (WNV) genetic diversity, evaluated using genetic alignment of complete genomic sequences. GenBank accession numbers are indicated on the tree branches of each virus; the first two or three letters stand for the country or the USA state reporting WNV (It = Italy, Sp = Spain, Mo = Morocco, Fr = France, Ken = Kenya, Rus = Russia, Tu = Tunisia, Hu = Hungary, Ro = Romania, Arg = Argentina, Tx = Texas, NY = New York, Is = Israel, Ind = India, Eg = Egypt, Kun = Kunjin Australia, SA = South Africa, Ug=Uganda, Ser = Serbia, and Gr = Greece) and the numbers indicate the year of isolation (96 = 1996, 10 = 2010). Japanese encephalitis virus (JEV), a closely related flavivirus, was used as an outgroup. The rooted phylogenetic tree was constructed using neighbor-joining with Jukes-Cantor parameter distances (scale bar) in MEGA (MEGA software, version 5.2 [25]). A bootstrapped confidence interval (1,000 replicates) and a confidence probability value based on the standard error test were also calculated using MEGA. The WNV strains responsible for recent human or equine outbreaks are underlined. The complete sequences of the most recent Romanian and Russian lineage 2 variants are not available, but at least two introduction events of lineage 2 strains have occurred in Europe: divergent lineage 2 strains have been observed in Romania/Russia and Hungary/Greece/Italy/Serbia/Austria [26].