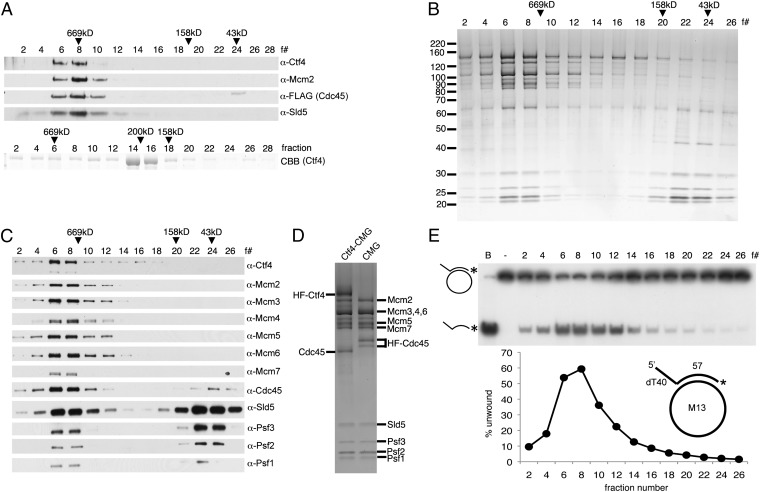
Fig. 1.
hCtf4 forms a stable complex with the hCMG complex. (A) Purified hCMG (5 pmol) (HF tag on Cdc45) and untagged hCtf4 (15 pmol, as a dimer) were mixed with anti-FLAG M2 affinity gel. (Upper) Bound proteins were eluted and subjected to glycerol gradient sedimentation. (Lower) As a control, untagged hCtf4 was sedimented separately. CBB, Coomassie brilliant blue. The hCtf4–CMG complex was purified as described in SI Materials and Methods, and each fraction (5 μL) was separated electrophoretically through a 4–20% (wt/vol) polyacrylamide gradient gel (Invitrogen), followed by silver staining (B) or Western blot analysis (C) against all 12 subunits of the hCtf4–CMG complex. The peak positions of protein markers (GE Healthcare Life Sciences) after sedimentation are indicated above fractions (669 kDa, thyroglobulin; 158 kDa, aldolase; 43 kDa, ovalbumin). (D) hCtf4–CMG and hCMG were loaded side by side onto a 4–20% (wt/vol) gel and silver-stained. (E) Helicase activity was measured across the hCtf4–CMG glycerol gradient (0.5-μL fractions), and the substrate unwound (%) was calculated and is shown in the graph. The structure of the helicase substrate containing a 57-mer duplex region and a 5′-dT40 tail (M13 annealed to labeled oligonucleotide no. 2) is shown in the graph. See Table S1 for description of oligonucleotides in this study. *, Location of 32P in substrate. B, boiled substrate.