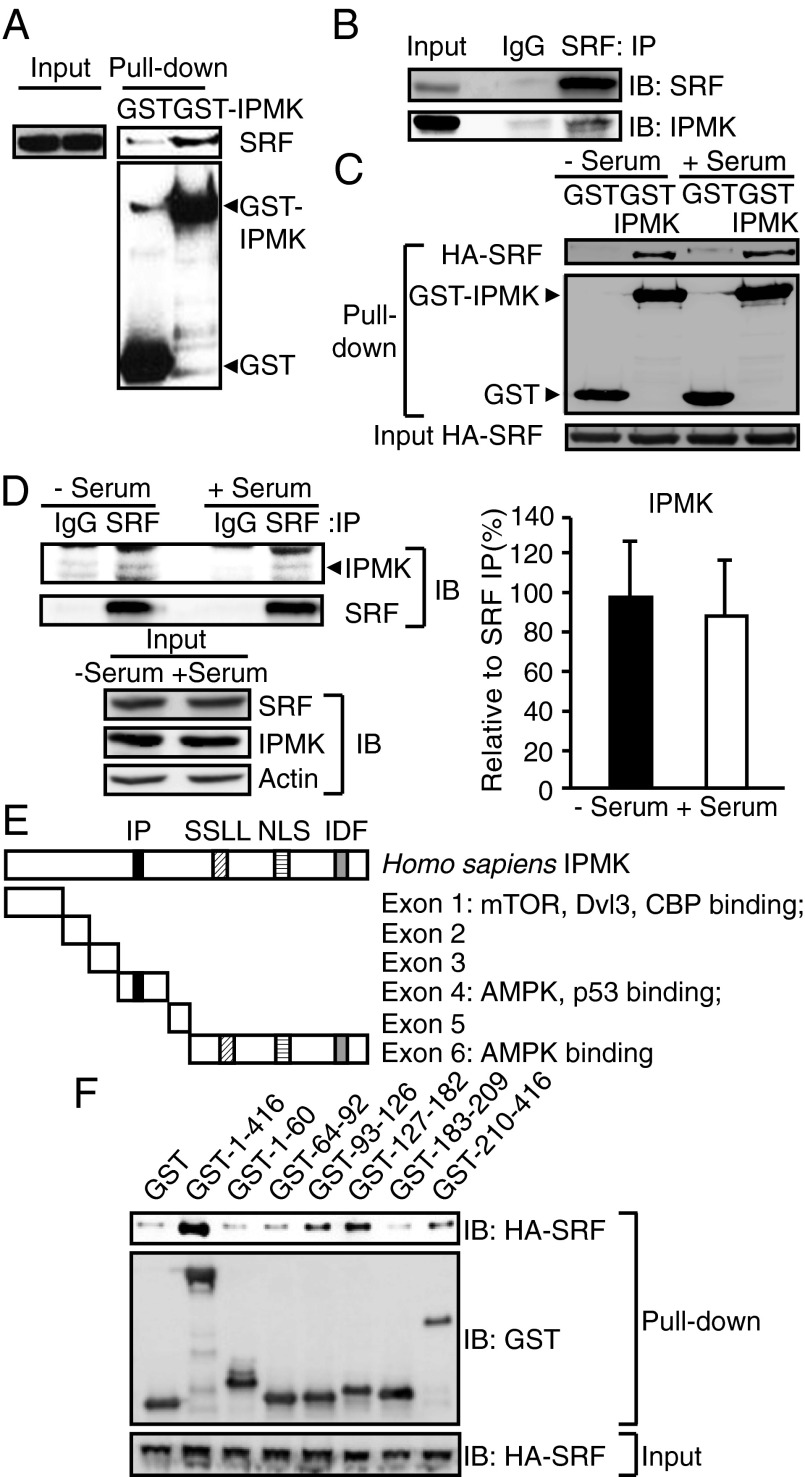
Fig. 3.
IPMK physically interacts with SRF. (A) Identification of SRF using GST-IPMK pull-down. GST or GST-IPMK was transfected in HEK293T cells followed by pull-down and immunoblot analysis for endogenous SRF. (B) Immunoprecipitation of IPMK and SRF proteins. MEF lysates were used for immunoprecipitaion (IP) against control IgG or SRF antibody to determine the binding between endogenous IPMK and SRF. (C) HA-SRF and GST-IPMK were cotransfected in HEK293T cells that were deprived of serum for 12 h and stimulated with 10% FBS for 30 min. GST-IPMK pull-down and immunoblotting were conducted. (D) SRF immunoprecipitates were prepared from MEFs that were deprived of serum for 12 h and stimulated with 10% FBS for 30 min. IPMK and SRF levels were analyzed by immunoblotting. Quantification of IPMK level normalized to coimmunoprecipitated SRF is presented as a graph. Bars represent mean ± SD (n = 4). (E) A schematic diagram of IPMK exon fragments with the numbers of amino acid sequences used for binding studies. IPMK-binding proteins were indicated. Key domains for inositol binding (IP), kinase activity (SSLL and IDF), and nuclear localization signal (NLS) are shown. (F) Mapping of binding region of IPMK responsible for SRF interaction. GST, GST-IPMK, or GST-IPMK exon fragments (exon 1, 1–63; exon 2, 64–92; exon 3, 93–124; exon 4, 125–182; exon 5, 183–208; and exon 6, 209–416) were pulled-down from HEK293T cells cotransfected with HA-SRF. SRF levels in IPMK pull-down were determined by immunoblotting.