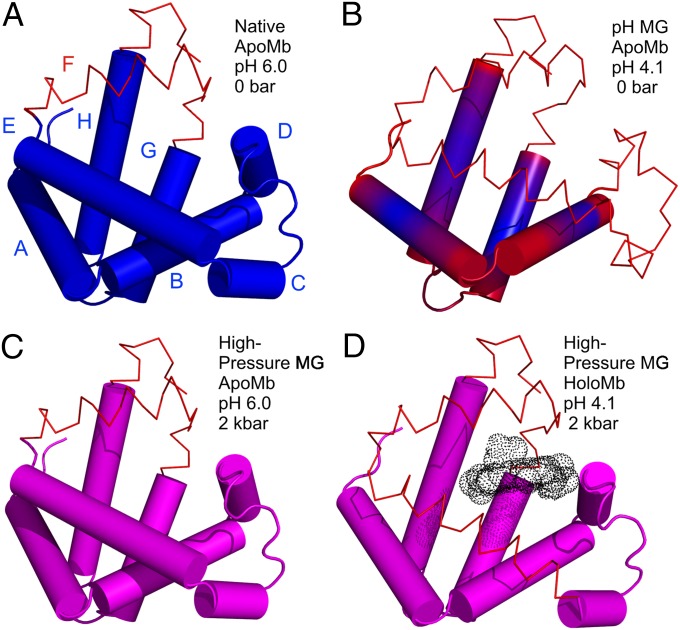
Fig. 5.
Models for the structure and dynamics of partially folded states of myoglobin. Each model is based on the crystal structure of holoMb; sequences that retain helical content are shown as cylinders; segments that have some fraction unfolded are shown in wire representation. (A and B) Models of native apoMb and the pH 4.1 MG state, respectively, based on solution NMR (42, 51). Regions undergoing conformational exchange are colored red, and more rigid regions are colored blue. The structure of native apoMb is similar to that of holoMb, but with localized unfolding in the F helix and portions of the G and H helices. In B, the conformational exchange implied by the gradient of color in the helices represents a gradient in the population of nonhelical states, increasing toward the helix termini, as inferred from 13Cα chemical shifts (42). (C and D) Models of the high-pressure MG of apoMb and holoMb, based on SDSL, CD, and NMR. Helices with a fluctuating tertiary structure are colored magenta. (C) The pressure-populated MG of apoMb at pH 6.0, 2 kbar, contains the full complement of native-state secondary structure according to CD, but line broadening in NMR and the spectral shifts in EPR indicate a fluctuating tertiary fold. (D) The pressure-populated MG of holoMb at pH 4.1 and 2 kbar experiences dynamic disorder in some fraction of the E and F helices, and a fluctuating tertiary fold in the remaining helices.