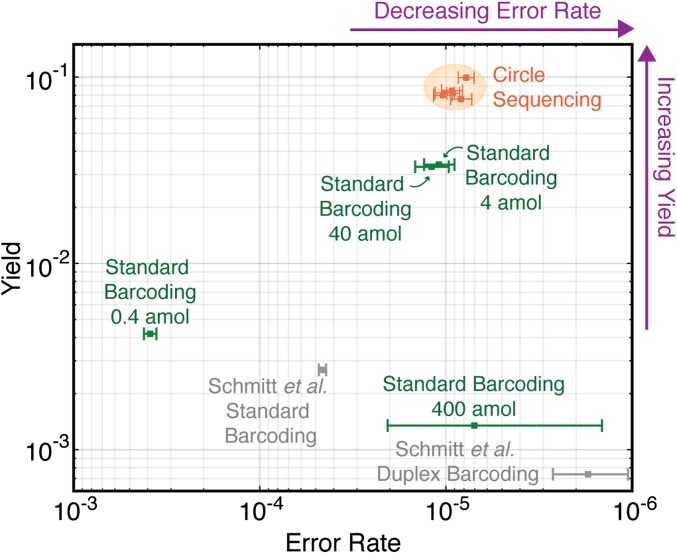
Fig. 4.
Comparison of overall yield and error rate for all error-correction methods. The yeast genome was sequenced with standard barcoding (green points) or circle sequencing (orange points) while varying input DNA concentration and/or reads produced. Error rate (x axis) is defined as the fraction of consensus bases that differ from the reference sequence. Yield (y axis) is the total number of consensus bases produced divided by the raw number of bases sequenced. Circle sequencing produces consistent error rates and yields across a range of experimental conditions (orange shading). Standard barcoding produces highly variable error rates and yields. Another library discussed in the text, from the M13mp2 phage genome generated in ref. 8, was also analyzed (gray points).