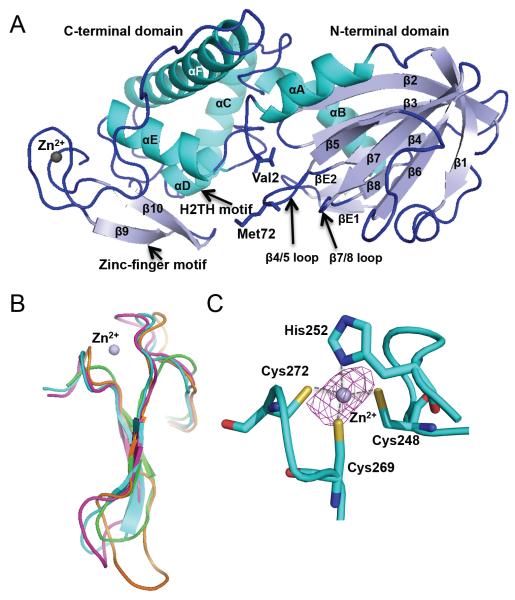
Figure 1.
Overall structure of Mimivirus Nei2. (A) Ribbon diagram of MvNei2. The secondary structure elements are as follows: αA (3–15), β1 (21–26), β2 (42–48), β3 (51–57), β4 (64–68), β5 (75–79), β6 (89–94), β7 (97–103), β8 (107–110), βE1 (117–119), βE2 (124–126), αB (127–140), αC (148–163), αD (167–173), αE (182–193), αF (205–232), β9 (255–259), and β10 (264–268). Helices are shown in cyan and β-strands in pale blue. The zinc metal ion is shown as a grey sphere. βE1 and 2 are unique to MvNei2 and are located before helix αB. (B) Superposition of the zinc/zincless-finger motifs from MvNei2 (cyan), MmuNeil3 (fuschia, PDB code 3W0F [16]), MvNei1 (orange, PDB code 3A46 [27]) and human NEIL1 (green, PDB code 1TDH [29]). The superposition was performed in COOT [41] using secondary-structure matching (SSM) of all residues in the enzymes. (C) Close-up view of the coordination of the zinc ion by His252, Cys248, Cys262, and Cys272. An anomalous difference Fourier map (magenta mesh) contoured at 3σ is displayed around the zinc ion.