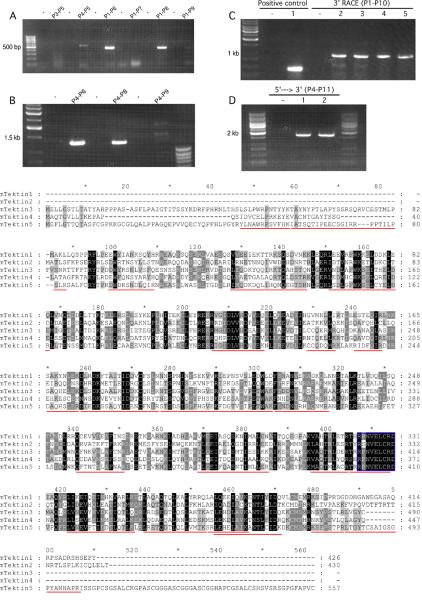
Figure 3.
Utilization of PCR and 3'-RACE to clone the testicular Tekt5 cDNA. The primers used in the molecular cloning procedure are labeled in Fig. 2A, and listed in Table 1. (A): Primer P4 was designed from the 5'-UTR region described for Tekt5 sequences present in the NCBI database. Primer P3 was based upon genomic sequence upstream from this region. P4 produced the expected size of product with P5 and was verified by DNA sequencing. P6, P7, P8, P9 are 3' corresponding primers for XM_908332, XM_980627, BC115969, and XM_156349, respectively. P6 and P8 produced the expected sizes of PCR products with P1. (B): Attempts to amplify longer PCR products with P4–P6 and P4–P8 were successful. The identities of the PCR products were verified by sequencing. (C): The 3'-UTR of Tekt5 was characterized by 3'-RACE. A positive control encompassing part of the Tekt5 open reading frame, (lane 1), was included to confirm that the RACE reverse transcription was successful. Gradient PCR reactions at different annealing temperatures (lanes 2–5 are 55°C, 57.5°C, 60°C, and 62.5°C, respectively) were used to optimize the RACE conditions. A predominant PCR product was seen at about 1 kb. (D): P11 was located in the 3'-UTR of Tekt5 RACE results. P4–P11 amplified the Tekt5 cDNA sequence from 5'-UTR to 3'-UTR (lanes 1–2 are 49°C, 52°C). The sequencing results showed that our Tekt5 cDNA sequence is similar to the computer-predicted sequence XM_908332, but that our Tekt5 cDNA sequence includes a complete polyadenylation signal (AATAAA) and a poly-A tail. Note: All PCR products were verified by nucleotide sequencing. The no-template control PCR reaction used water in place of the sample. (E): alignment of encoded Tektin 5 with other known Tektins. The peptide sequences identified by mass spectroscopy are underlined in red and the canonical tektin domain is outlined with a blue box.