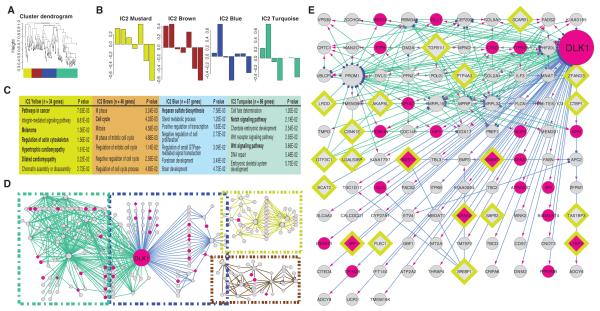
Fig. 4.
Combined topological overlap–based clustering and dynamic Bayesian network construction links Wnt1 signaling with changes in dementia-related genes. (A and B) WGCNA clustering of ICM2 genes: Wnt1-stimulated expression time courses for the genes that compose the ICM2 module were averaged and then subjected to TOM-WGCNA–based clustering (A). This produced four submodules (Mustard, Brown, Blue, and Turquoise). (B) Submodule eigengenes, where singular value decomposition was used to extract a characteristic first principal component eigengene for each submodule. The y axis is eigengene expression. (C) GO analysis reveals functional uniqueness of individual submodules. (D and E) Dynamic Bayesian network (DBN) depicting causal relationships, within each module. (D) Overview of the DBN-based causality network. Edge color codes the original submodule. Node color indicates those genes identified by DTW analysis (magenta; n = 23). Outlined diamonds denote those genes whose expression was increased in the brains of Alzheimer’s patients (n = 20) (70). DLK1 forms the primary hub in this network. [Note: SORT1, like DLK1, is a binding partner for PGRN (73).] (E) A more detailed view of the DLK1 hub and its associated genes, revealing a significant overlap (hypergeometric P ≤ 0.001) with DTW-identified genes and a strong enrichment for genes with increased message in AD brains. Transcriptional drivers (blue dots), targets (magenta arrowheads). (See fig. S11 for scalable version of this figure.)