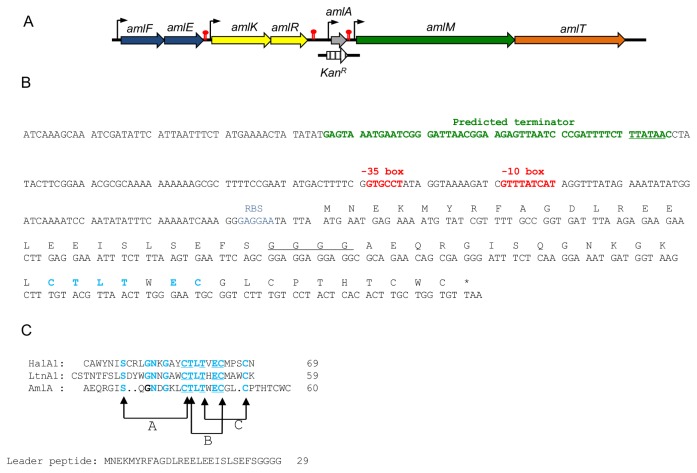
Figure 1. Characterization of the amylolysin gene cluster.
(A). The gene coding for the amylolysin prepeptide (amlA) is colored in grey. The other genes are marked with the following patterns: immunity (amlEF) in blue, regulation (amlKR) in yellow, modification (amlM) in green and transport in orange. Stripes arrow corresponds to the Kan construct used for amlA disruption. Putative promoters and terminators are indicated by vertical arrows and red symbol, respectively. (B) Primary structure of amlA gene. Predicted -10box and -35box are in red, ribosome binding site (RBS) is in blue and predicted terminator of amlKR cluster is in green. The four-glycine stretch is underlined and the highly conserved motif CTLTXEC is in bold blue. (C) Amino acid alignment of AmlA with LanA1 peptides of the two-peptide lantibiotics haloduracin (HalA1, DAB04173) and lacticin 3147 (LtnA1, O87236). Conserved amino acids are in bold blue; those forming the CTLTXEC motif are in bold and underlined. The thioether bridging patterns represents that of HalA1.