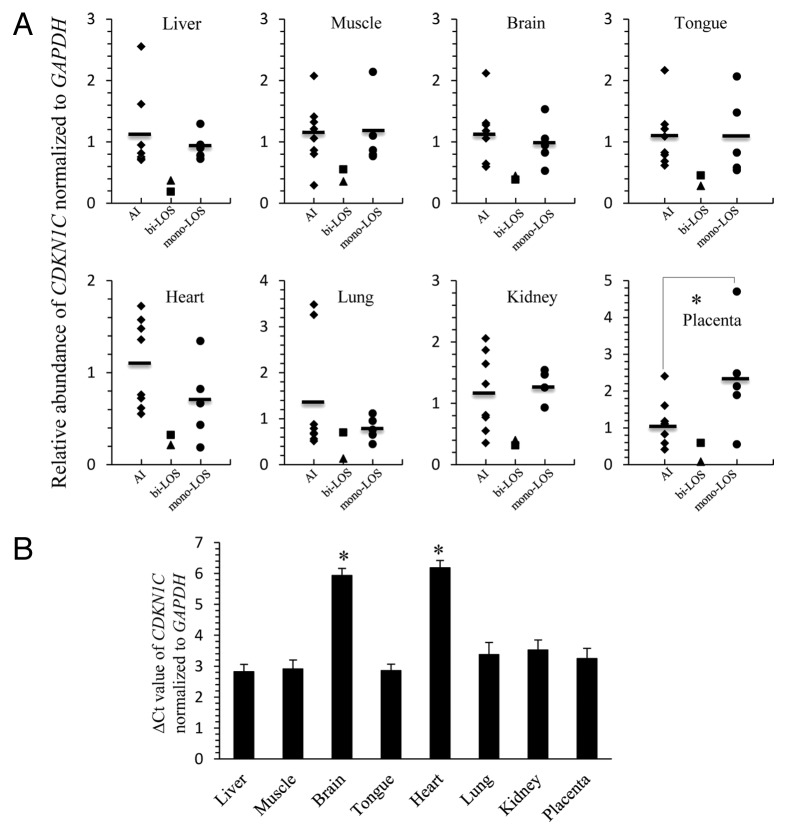
Figure 4. LOS fetuses with biallelic expression of KCNQ1OT1 show downregulation of CDKN1C. (A) The CDKN1C RNA was determined by quantitative RT- PCR in several tissues from eight AI fetuses (diamonds; AI), two LOS fetuses with biallelic expression of KCNQ1OT1 (triangle = ART-J835LOS; square = ART-J489ALOS; bi-LOS), and five LOS fetuses with correct imprinting of KCNQ1OT1 (circles; mono-LOS). The short line among the diamonds and circles represents the average level of the individuals. CDKN1C level was normalized to the expression of GAPDH. Note that for each tissue analyzed CDKN1C level in ART-J835LOS is at least 2-fold lower than the average level found in AI fetuses and LOS fetuses with correct imprinting of KCNQ1OT1. It should be also noted that lung from ART-J489ALOS which had correct imprinting of KCNQ1OT1 (Fig. 3) exhibits the comparative CDKN1C level with controls. (B) The threshold cycle (Ct) of CDKN1C was normalized to the reference gene GAPDH in each tissue from the eight AI fetuses. The data are expressed as mean ± SEM. *p < 0.05 between the brain, heart and other tissues analyzed. LOS, large offspring syndrome.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.