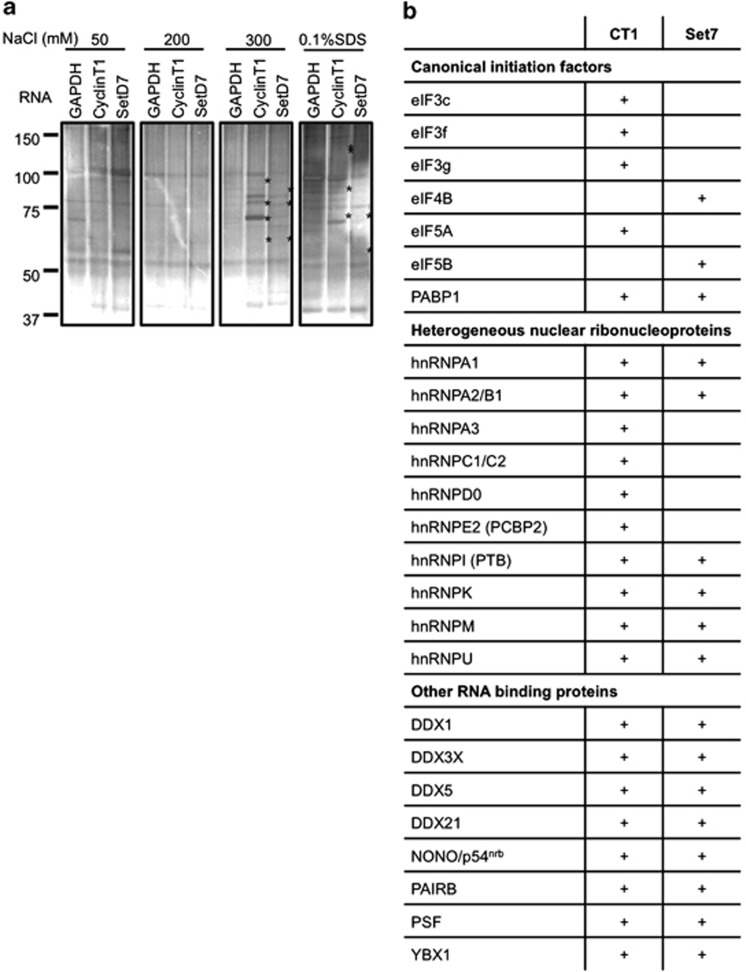
Figure 3.
IRES RNA affinity analysis. (a) In all, 50 μg of biotin-tagged cyclin T1 and SETD7 5' UTRs or 300 nt RNA of GAPDH coding region was used as bait to purify IRES-binding proteins from HeLa cell lysate in a binding buffer (10 mM Tris pH 7.5, 25 mM KCl, 2 mM MgCl2, 0.02% Tween-20, 1 mM ATP, 1 μg/ml yeast tRNA, 1 μg/ml heparin, RNase inhibitor, 1 × protease inhibitor) for 60 min at 4 °C. Streptavidin-conjugated magnetic beads (Invitrogen) were added and incubated for a further 30 min. The bound complexes were washed with buffers that contained increasing salt concentration and eluted samples were applied to SDS-PAGE and stained with colloidal Coomassie blue and silver. Proteins that were specifically eluted from the cyclin T1 or SETD7IRESs, but not the GAPDH are marked (*). (b) Proteins bound to each RNA were identified firstly via LC-MS/MS mass spectrometry (Supplementary Figures S2A and B) and then via immunoblotting analysis (Supplementary Figure S2C). The table shows proteins, which were identified as binding to either or both of the cyclin T1 and SETD7 IRESs, whereby a plus indicates binding, in either the mass spectrometry data, the immunoblotting data or both. The proteins included in the table are those found to be both IRES binding and PTB associated