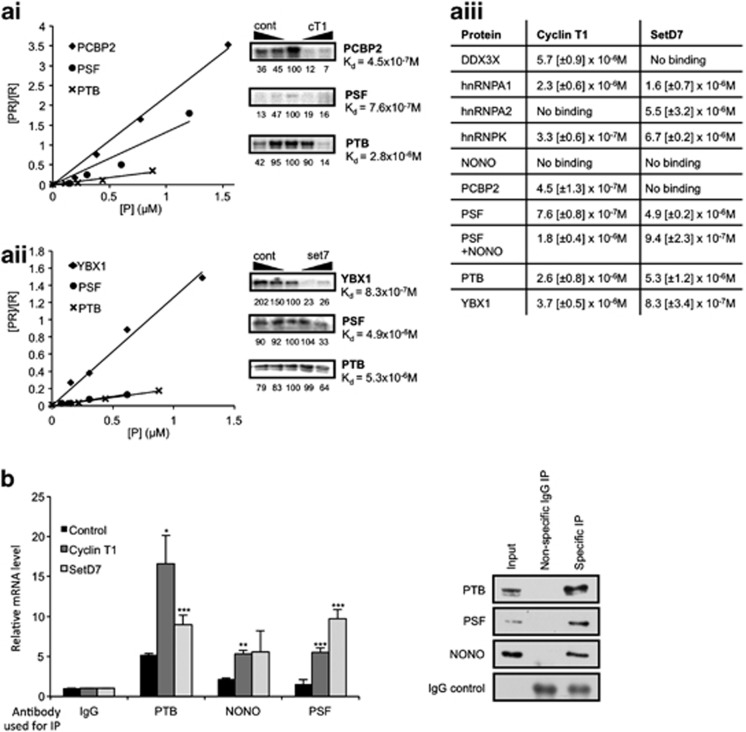
Figure 4.
Members of the PTB-containing complex bind apoptotic IRES RNA in vitro and in vivo. Binding of the identified complex members to the apoptotic IRES RNA was validated by a number of methods. (a) UV-crosslinking (right panels) and filter-binding assays (left panels) using 32P-labelled cyclin T1 or SETD7 5' UTR. (ai) 32P-CTP-labelled cyclin T1 and rePTB, rePSF and rePCBP2 or (aii) 32P-CTP-labelled SETD7 RNA with rePTB, rePSF and reYBX1 were used in UV-crosslinking and filter-binding assays. Specificity of the binding was demonstrated by increasing the amount of cold competitor IRES RNA (right panel), or nonspecific control RNA (left panel). The UV-crosslinked signal was quantified relative to the central no competitor RNA lane, which was set to a 100. (aiii) Dissociation constants for each protein/RNA combination were calculated in the filter-binding assay. A constant level of the 32P-labelled probe was used over a range of molarity of each recombinant protein. S.D. is shown in square brackets, and the data are the average of three repeats. (b) Binding of endogenous PTB, PSF or NONO/p54nrb to the IRES RNA was shown using RNA-IP with antibodies against the proteins indicated, followed by RT-qPCR using primers specific for cyclin T1, SETD7 or control RNA. 15 cm confluent plates of HeLa cells were lysed in lysis buffer (20 mM HEPES pH7.2, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, RNAsin 400 U/ml, 0.5% Triton X-100 and protease inhibitors), incubated for 5 min at 4 °C and centrifuged to pellet the nuclei. The post-nuclear extract was incubated with PTB, PSF or NONO/p54nrb antibodies or IgG (control) coated protein A beads for 1 h at 4 °C. Beads were washed 4 × 30 min with buffer A (20 mM Hepes pH 7.2, 200 mM NaCl, 5 mM MgCl2, 0.5% Triton X-100, 1 mM DTT) and 2 × with buffer B (50 mM Tris pH 7.4, 150 mM NaCl, 1 mM MgCl2 and 0.05% NP40). Beads containing protein-RNA complexes were isolated and re-suspended in 200 μl buffer B and proteinase K treated for 45 min at 55 °C. The RNA was isolated by phenol/chloroform extraction and quantified by RT-qPCR using primers specific for cyclin T1, SETD7 or control RNA. Results are shown relative to a nonspecific IgG immunoprecipitation. Western blot of the IPs using antibodies against proteins indicated is shown on the right. Significance (*P<0.05, **P<0.01 or ***P<0.005) was calculated using an unpaired two-tailed Student's t-test (n=3), error bars represent S.D.