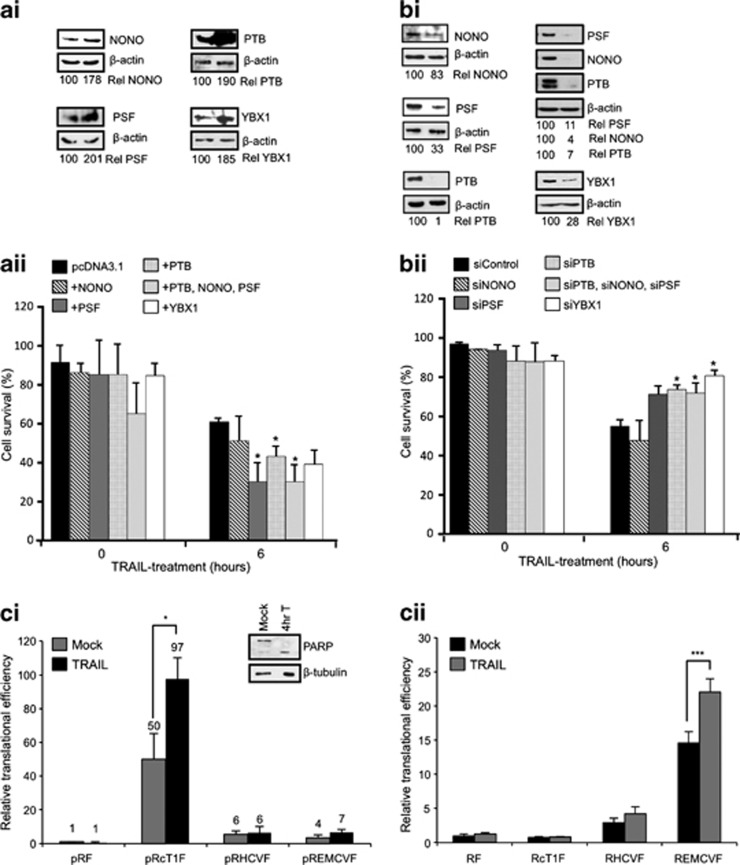
Figure 6.
Altered expression of PTB-binding proteins controls apoptotic rates. (a) Overexpression of PTB1, NONO/p54nrb, PSF either individually or together enhances TRAIL-induced apoptosis. (ai) MCF7 cells were transfected with pcDNA3.1, pcDNA3.1-PTB1, pcDNA3.1-NONO/p54nrb, pcDNA3.1-PSF, pcDNA3.1-YBX1 plasmids and subjected to 6-h TRAIL treatment. The overexpression was assessed by western blot using antibodies against the proteins indicated. Immunoblots were quantified relative to β-actin levels and expressed as a percent of levels in the control cells. (aii) FACS analysis of MCF7 cells overexpressing the proteins above. After TRAIL treatment, cells were stained with Annexin V-FITC and propidium iodide and subjected to FACS analysis. Significance (*P<0.05) was calculated using an unpaired two-tailed Student's t-test (n=3) relative to the control. (b) Depletion of endogenous YBX1, PTB, NONO/p54nrb and PSF individually or in combination protected cells against TRAIL-mediated apoptosis. (bi) Western blot of siRNA treated MCF7 cells using antibodies indicated. Immunoblots were quantified relative to β-actin levels and expressed as a percent of levels in the control cells. (bii) FACS analysis of siRNA treated MCF7 cells treated as in aii. Significance (*P<0.05) was calculated using a paired two-tailed Student's t-test (n=3) relative to the control. (ci) MCF7 cells were transfected via nucleofection with dicistronic pRIRESF plasmid, incubated for 48 h before mock or TRAIL treatment for 4 h. Both protein and RNA were harvested. Firefly luciferase counts were normalized to total protein, and luciferase RNA levels were normalized to GAPDH RNA. The translation efficiency is expressed as Firefly luciferase activity/mRNA level. Data are presented relative to this value for pRF in the mock condition. Western blot with αPARP was performed to monitor the degree of apoptosis. Significance (*P<0.05) was calculated using an unpaired two-tailed Student's t-test (n=3), error bars represent S.D. (cii) In all, 1 μg of capped and polyadenylated mRNA was transfected into MCF7 cells, incubated for 2 h before mock or TRAIL-treatment for a further 4 h. Cells were harvested and assayed for both Renilla and Firefly luciferase. Firefly luciferase counts were normalized to Renilla luciferase counts and are presented relative to RF in the mock condition. Significance (***P<0.01) was calculated using an unpaired two-tailed Student's t-test (n=5), error bars represent S.D.