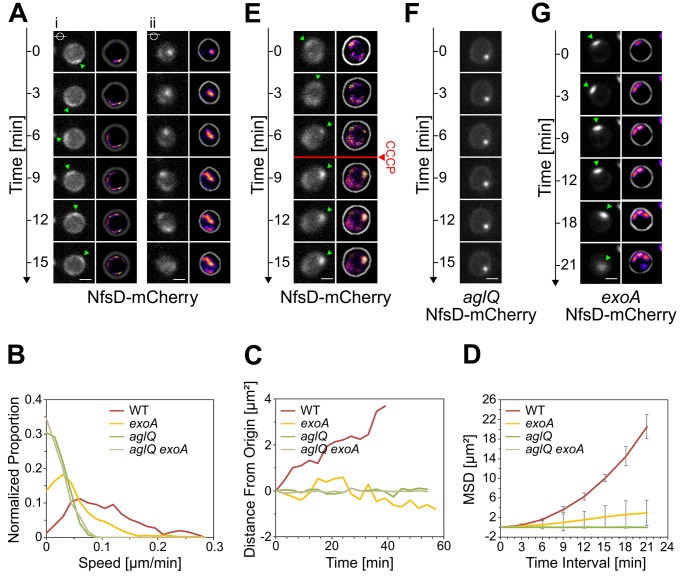
Figure 4. The AglRQS motor rotates the Nfs complex around the spore surface.
(A) NfsD-mCherry moves in orbital trajectories around the surface of 4-h-old spores. Observation of dynamic NfsD-mCherry clusters at different focal planes, middle (i) and top (ii) sections of a spore. For each time lapse, trajectories were computed by summing the different time points in consecutive frames and shown in “fire” colors to the right. Scale bar = 1 µm. (B) Instantaneous speed histogram of tracked NfsD-mCherry clusters over time in WT spores and in the different mutants. (C) Distance from the origin of NfsD-mCherry clusters in WT spores and different mutants. (D) Mean square displacement (MSD) of NfsD-mCherry clusters over time in WT spores and in the different mutants. (E) NfsD-mCherry rotation is abolished by CCCP. (F) Rotation of NfsD-mCherry requires AglQ. (G) Movement of NfsD-mCherry in the exoA mutant.