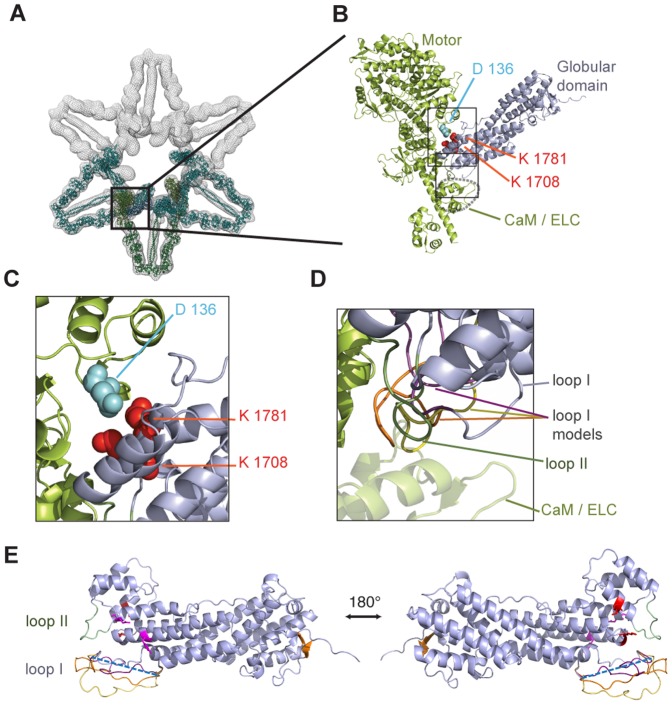
Figure 4. Autoinhibition of type V myosins by their globular domains involves multiple interactions sites.
(A) Docking of our high resolution Myo5a GD structure into a previously published 24 Å electron density map of the inhibited Myo5a motor [21] (PDB-ID of published model lacking the globular domain: 2DFS). Shown are three modeled myosin dimers in the EM density (meshed surface rendering) that are arranged in a flower-like fashion. The color scheme is as follows: blue, Myo5a GD; green, motor domain, lever arm, light chains, coiled coil domain; turquoise, neighboring dimers. (B) Close-up of (A), depicting the Myo5a head complex (green) and the fitted GD (blue). Residues previously reported to be required for autoinhibition [23] are shown as colored spheres. (C) Close-up of the upper rectangle in (B). (D) Close-up of the lower rectangle in (B). Loop I is disordered in the structural data. Depicted are three computed models for the flexible loop I, highlighted in yellow, orange, and magenta. (E) Atomic model of Myo5a. Missing amino acids of the flexible loop I GD are depicted as a blue dashed line (disordered loop I: 1640–1658), loop II in green (1787–1797), and amino acids important for interaction with the motor domain are highlighted in red (K1708, K1781). Residues important for Rab11a binding (Y1721 and Q1755) are shown in magenta and the beta-sheet is depicted in orange. Computed models for the flexible loop I were depicted in yellow, orange, and magenta (see also D). Figures were generated with the program Pymol.