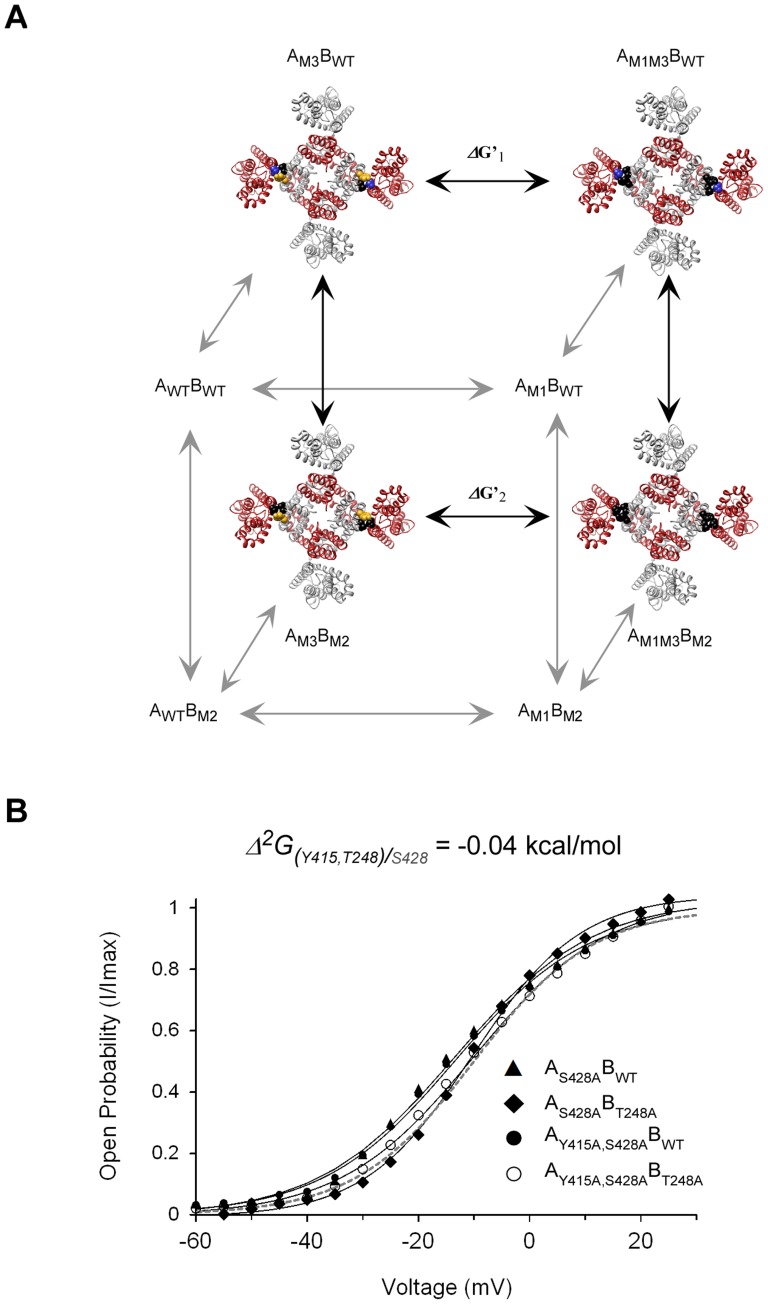
Figure 5. The upper interface T248, Y415 and S428 residue triad forms a network of coupled interactions during activation gating.
A) A thermodynamic cubic construct was used to measure mutual three-dimensional coupling (Δ 3 G(i,j,k)) between the three upper interface residues. Δ 3 G is calculated by subtracting the coupling free energy between any residue pair in the presence and absence of the native third residue (front and back faces of the cube, respectively). For clarity, only the channel proteins of the back face are explicitly represented. The front face proteins are described in the legend to Fig. 4A. B) Voltage-activation curves for four channel proteins comprising the thermodynamic double-mutant cycle measuring the coupling free energy between the upper interface T248 and Y415 (i,j) residues on the background of the mutated S428 residue (k). The same cycle, only in the presence of the native third position, is considered in Fig. 4B (front face of the cubic construct shown in (A)).