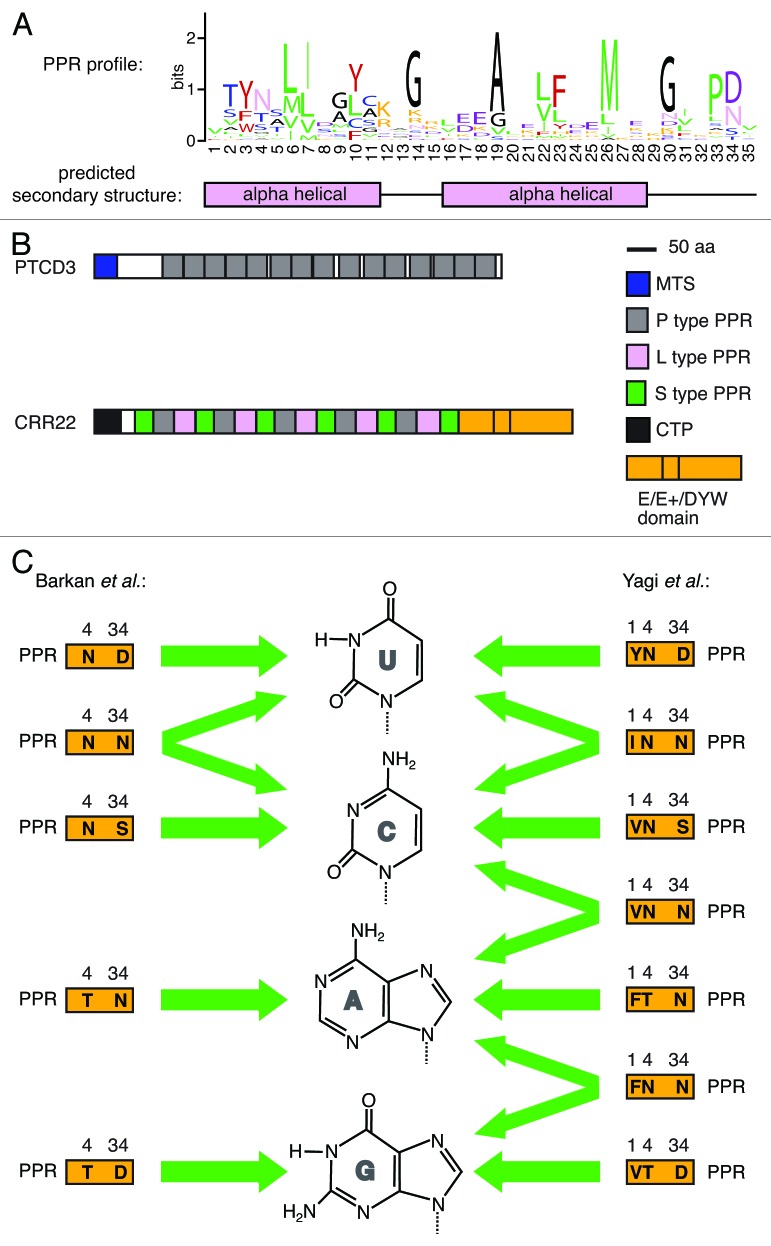
Figure 1. Pentatricopeptide repeats. (A) A sequence logo illustrating the characteristic amino acid composition of PPR sequences. The logo was derived from 14,466 PPRs found in the PROSITE PPR entry (PDOC51375) using WebLogo.83 These sequences are derived from the following taxonomic groups: 86% plants, 5.7% fungi, 4.3% animals, 1.8% algae, 1% trypanosomes and 1.2% others. Amino acids are color-coded according to the physiochemical properties of their side chains: small (A, G) in black, nucleophilic (C, S, T) in blue, hydrophobic (I, L, V, M, P) in green, aromatic (F, W, Y) in red, acidic (D, E) in purple, amides (Q, N) in pink and basic (H, K, R) in orange. Regions of α-helical structure are shown below. Amino acids are numbered based on the Pfam model, which functions as a minimal unit.54 Residue 34 is also defined as ii according to Kobayashi et al.,54 while the numbering scheme used by Fujii et al.53 is shifted to the N terminus by two amino acids such that amino acids 1, 4 and 34 in the Pfam model are annotated as 3, 6 and 1, respectively. (B) Schematic structures of a typical P class PPR protein, human PTCD384 and a typical PLS class PPR protein, Arabidopsis CRR22.85 PPRs, mitochondrial targeting sequence (MTS), chloroplast targeting peptide (CTP) and the E/E+/DYW domain, often associated with editing PPR proteins, are highlighted. (C) The recognition code of PPRs for RNA bases. Only representative predictions by Yagi et al. are shown; for a full list, refer to the original research paper.57

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.