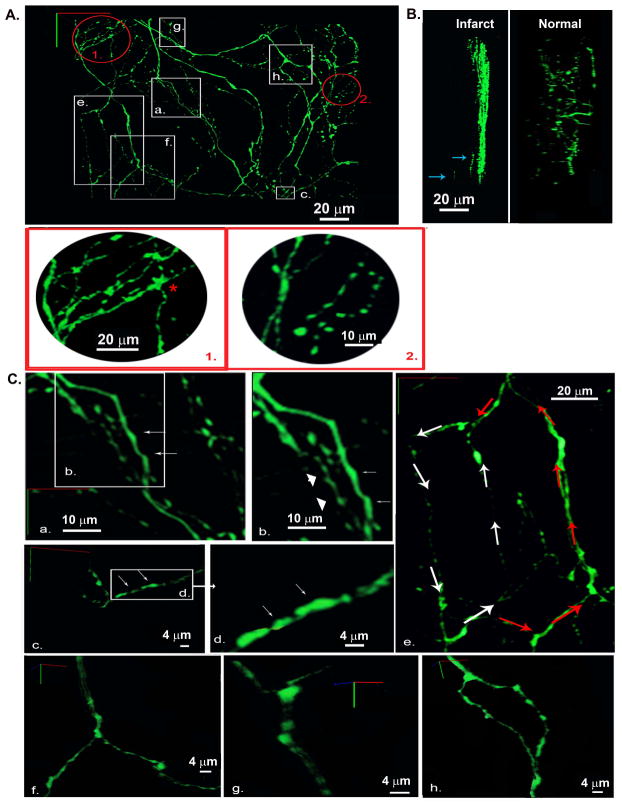
Figure 7.
Three-dimensional reconstruction of the local sympathetic neurite network within the epicardial border zone of a 2-week-old myocardial infarction in a living, adult hDβH-EGFP heart. A, 3-D rendering of the sympathetic arbor. All sympathetic axons (labeled with EGFP) in a 230 μm by 250 μm by 50 μm segment were imaged by TPLSM. A montage of 9 (3 × 3) overlapping image stacks provided the dataset for reconstruction. Images (512 × 512) were collected at a voxel size of 0.2 μm × 0.2 μm × 0.5 μm with 12-bit resolution. Red-boxed regions 1 and 2 are magnified views of the circled regions 1 and 2 in A demonstrating locally increased axon density. B, 3-D renderings at a 90°-rotation about the y axis of 3-D data sets obtained from an infarcted and a non-infarcted hDβH-EGFP heart. Arrows denote axonal fragments at depth. C, Magnified views of the boxed regions labeled a through h in A. a and b, axons with smooth contours (arrows) and with periodic swellings (arrowheads); c and d, eccentric boutons (arrows); e, two axonal loops with different circumferential length; f and g, branching of dual-axon bundles while their constituent axons do not branch; h, bifurcation where the parent axon branches into childs.