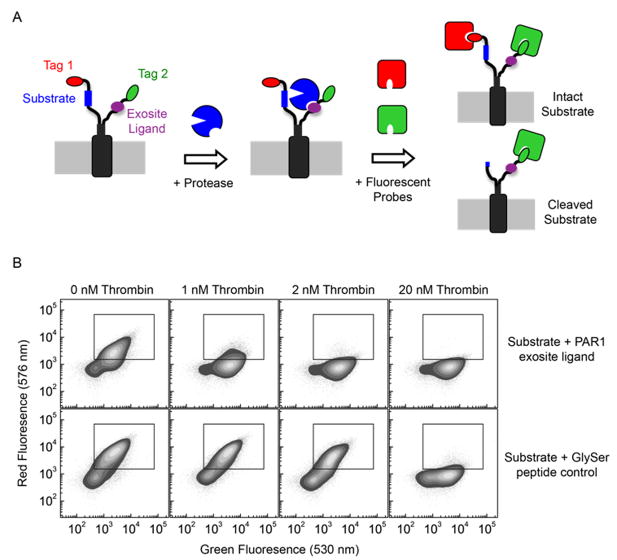
Figure 1. Method for the discovery of exosite ligands using eCLiPS.
A. Using a fixed substrate recombinantly fused to the N-terminus of the bacterial display scaffold, a library of candidate exosite binding peptides is constructed within the C-terminus and quantitatively screened for accelerated substrate cleavage using FACS based on a two-color labeling scheme. B. Flow cytometric analysis of cell populations displaying the PAR1 substrate and either the PAR1 exosite ligand or the control GlySer peptide, following incubation with three different thrombin concentrations.